Genome Sequencing of Mekong Tiger Perch: Insights into Phylogeny and Conservation
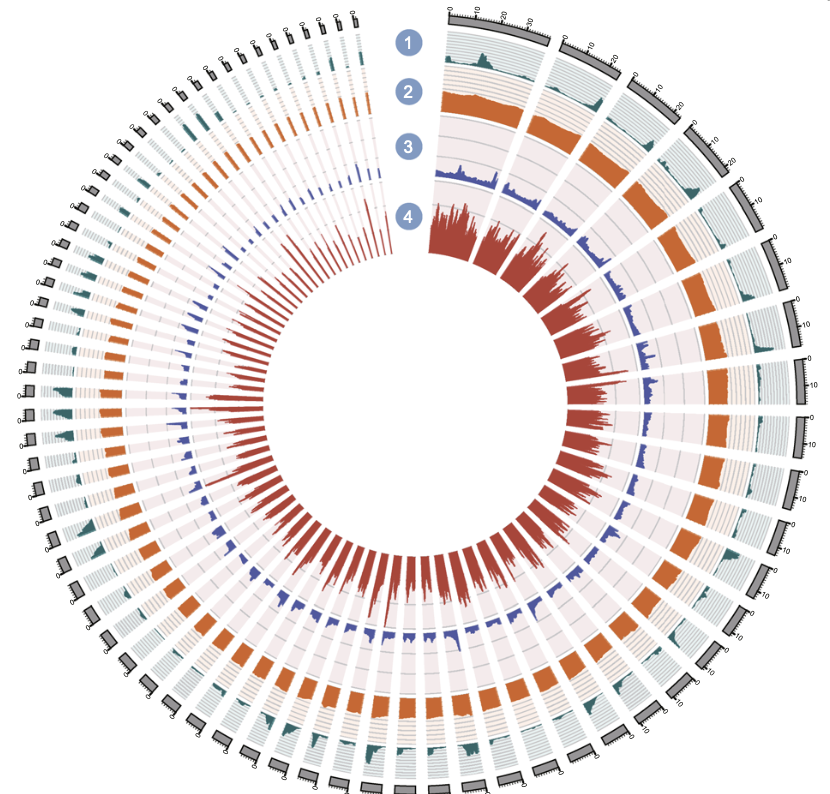
The Mekong tiger perch (Datnioides undecimradiatus) is a vulnerable freshwater fish native to the Mekong basin in Indochina. As an ornamental species with ecological significance, understanding its genetic makeup is crucial for conservation efforts. In our latest study, published in Scientific Reports, we present the first whole-genome sequence of the Mekong tiger perch, shedding light on its phylogenetic position and genetic adaptations.
Key Findings
-
High-Quality Genome Assembly:
Using stLFR co-barcoding and Oxford Nanopore sequencing, we generated a 595 Mb genome assembly, marking the first reference genome for the order Lobotiformes. -
Resolving Phylogenetic Uncertainty:
Phylogenetic analysis confirmed that Lobotiformes is more closely related to Sciaenidae than to Tetraodontiformes, resolving a long-standing debate in fish taxonomy. -
Pigment Development and Genetic Conservation:
We identified four key rate-limiting genes involved in pigment synthesis, which have been retained following fish-specific genome duplication events. -
Demographic History and Population Decline:
Our study estimated the effective population size of Mekong tiger perch, revealing a continuous reduction, potentially linked to the contraction of immune-related genes.
Reflections
This research represents a significant step in understanding the evolutionary history and conservation needs of Mekong tiger perch. By collaborating with experts in fish genomics, we successfully assembled and analyzed the genome, providing valuable insights into phylogenetics, pigmentation, and population dynamics. The findings not only contribute to taxonomy and evolutionary biology but also offer genomic resources for conservation strategies.
The full text of this study can be accessed online at Scientific Reports.