SLR-superscaffolder: A Novel Scaffolding Tool for Synthetic Long Reads
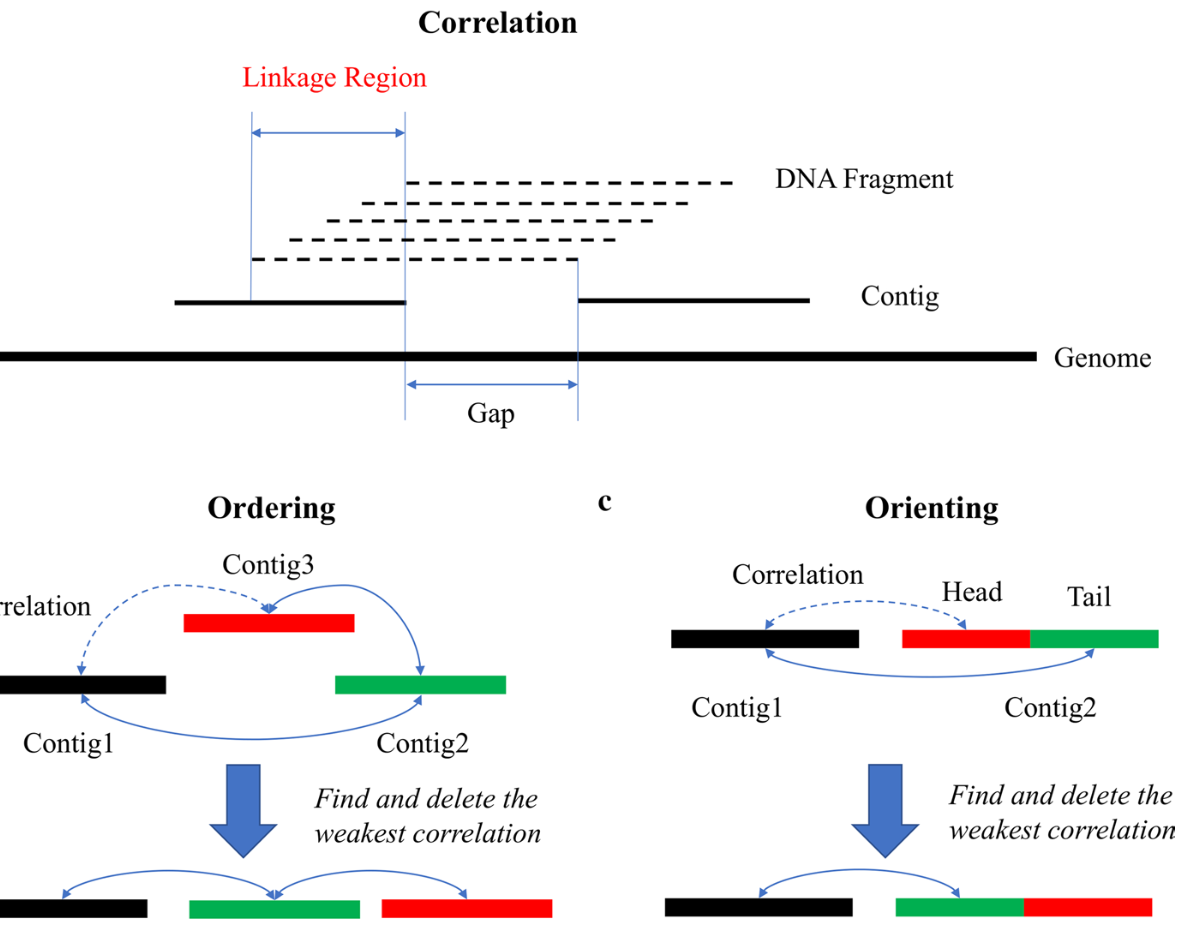
Genome assembly is a fundamental component of genomics research, enabling the reconstruction of complete genetic sequences from raw sequencing data. With the advancement of sequencing technologies, synthetic long reads (SLR) have emerged as an efficient strategy to improve genome assembly quality. In our latest study, published in BMC Bioinformatics, we introduce SLR-superscaffolder, a powerful standalone scaffolding tool designed to enhance assembly contiguity using co-barcoding and paired-end sequencing data.
Key Findings
-
Standalone Scaffolding for Synthetic Long Reads:
SLR-superscaffolder integrates co-barcoding and paired-end information to link draft assembly contigs into scaffolds, significantly improving genome assembly contiguity. -
Innovative Top-to-Bottom Approach:
Unlike previous scaffolding tools, our algorithm first constructs a global scaffold graph based on Jaccard Similarity, determining the order and orientation of contigs before performing local refinements. This hierarchical approach reduces errors and misassemblies that often arise in genome assembly. -
Robust Performance Compared to Existing Scaffolders:
We benchmarked SLR-superscaffolder against other scaffolding tools, including fragScaff, Architect, and ARKS, and demonstrated superior results—achieving longer scaffold NG50 values and fewer misassemblies. -
Application in Human Genome Assembly:
Our tool was applied to the human genome sequencing dataset (HG001), improving the scaffold NG50 1,349-fold, reinforcing its effectiveness for large-scale genome projects.
Reflections
Genome assembly algorithms are a crucial part of bioinformatics research. At BGI, particularly after 2014, our focus shifted toward developing sequencing technologies based on our proprietary platforms. However, during this period, long-read sequencing became increasingly favored for genome assembly due to its superior continuity and accuracy. From both an experimental and computational perspective, long-read sequencing simplifies genome assembly while producing higher-quality results.
Yet, at that time, most of our sequencing platforms were optimized for short-read sequencing, making genome assembly more challenging. Fortunately, we had developed single-tube long fragment read (stLFR) sequencing, a technique that functions like BAC library sequencing for short-read platforms. This approach provided an effective means of improving genome assembly through scaffolding. In this study, we utilized stLFR sequencing data for scaffolding, successfully enhancing genome assembly quality and continuity. This research is an important step forward in combining short-read sequencing with advanced assembly strategies.
The full text of this study can be accessed online at BMC Bioinformatics.