Symbiont-Screener: Efficient Separation of Host and Symbiont Sequences for Long-Read Metagenomics
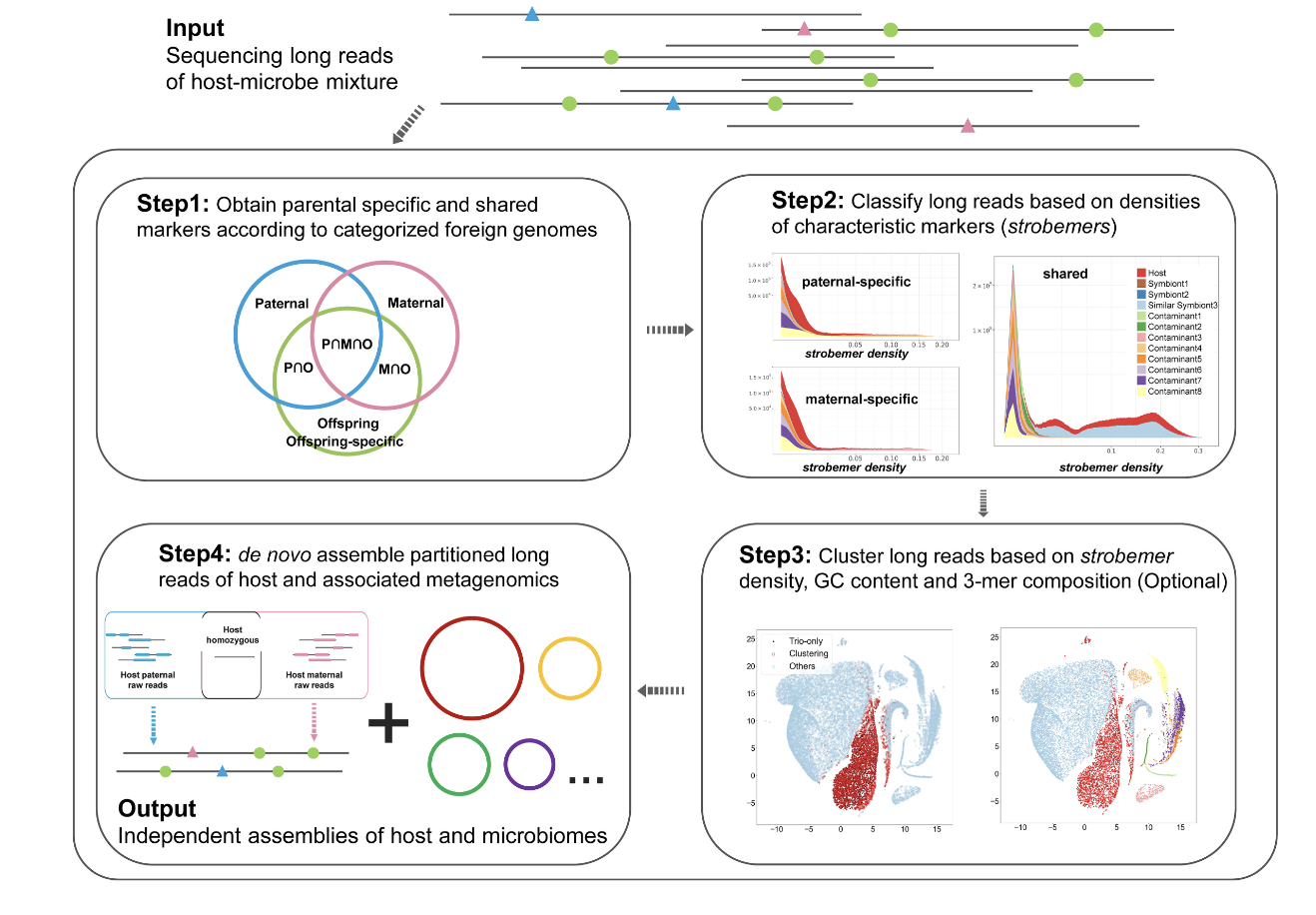
The phenomenon of symbiosis is widespread in nature, with biological samples often containing data from multiple species. In such cases, sequencing results are frequently mixed with data originating from unintended species, which we often term “contamination.” Removing this contamination to isolate host-specific sequences is critically important for high-quality genomic research. To address this challenge, our recent study introduces Symbiont-Screener, a reference-free computational tool designed to efficiently differentiate and separate sequences from different sources based on their unique sequence characteristics.
Key Findings
-
Reference-Free Approach for Host-Symbiont Separation:
Symbiont-Screener operates without relying on pre-existing reference genomes, enabling effective identification of host sequences in mixed samples. It utilizes trio-binning techniques and strobemers (error-tolerant genomic markers) to categorize long-read sequences according to their parental origins, GC content, and sequence composition. This ensures higher precision in host genome reconstruction and improved metagenomic profiling for associated microbiomes. -
Performance Across Simulated and Real Datasets:
Using both simulated datasets and real-world samples (e.g., algal-bacterial symbiotic systems), Symbiont-Screener demonstrated significant improvement in host genome assembly quality. Long-read sequencing technologies like PacBio HiFi and Oxford Nanopore were leveraged, with the purified host assemblies achieving chromosome-level contiguity while reducing contamination effectively. The remaining symbiont reads also facilitated more accurate profiling and assembly of associated microbial genomes. -
Implications for Symbiosis Research:
Beyond separating host and symbiont sequences, this tool serves as a foundational step for exploring the biological significance of symbiotic relationships. By enabling precise partitioning of sequences, Symbiont-Screener lays the groundwork for deeper investigations into the ecological and evolutionary mechanisms driving symbiosis.
Reflections
This project was led by Mengyang Xu, who not only served as the primary executor of the study but is also the team leader for software development at BGI-Qingdao. The innovative approach presented by Symbiont-Screener addresses a common challenge in genomic sequencing, where samples often include data from multiple species. Mengyang’s dedication to developing and testing this methodology has been pivotal to its success.
On a personal note, as someone involved in symbiosis-related projects, I find it fascinating to consider the broader biological significance of these intertwined relationships. While Symbiont-Screener focuses on separating host and symbiont sequences, this separation represents just the beginning of research into the reasons and mechanisms behind symbiosis itself. Understanding this phenomenon requires a deeper exploration into why symbiotic systems arise and persist in nature—a challenge that we are keen to tackle in the future.
Conclusion
Symbiont-Screener exemplifies the power of bioinformatics in solving complex challenges within metagenomics and symbiotic studies. Its ability to efficiently separate host sequences from symbionts and contaminants not only facilitates high-quality genomic assembly but also provides a novel perspective for investigating symbiotic relationships. As we advance this research, we look forward to uncovering more insights into the co-evolution and functional dynamics of symbiotic organisms.