Chromosome-Level Antarctic Krill Genome Unlocks Insights into Adaptations and Population Dynamics
Antarctic krill (Euphausia superba), Earth’s most abundant wild animal, plays a pivotal role in the Southern Ocean ecosystem. This species’ enormous biomass supports marine food webs and influences global biogeochemical cycles. In our recent study published in Cell, we present the chromosome-level genome assembly of Antarctic krill—the largest animal genome ever sequenced at 48.01 Gb. Our findings provide a unique lens into the genetic adaptations of krill to the harsh and highly seasonal Antarctic environment, as well as their population dynamics.
Key Findings
-
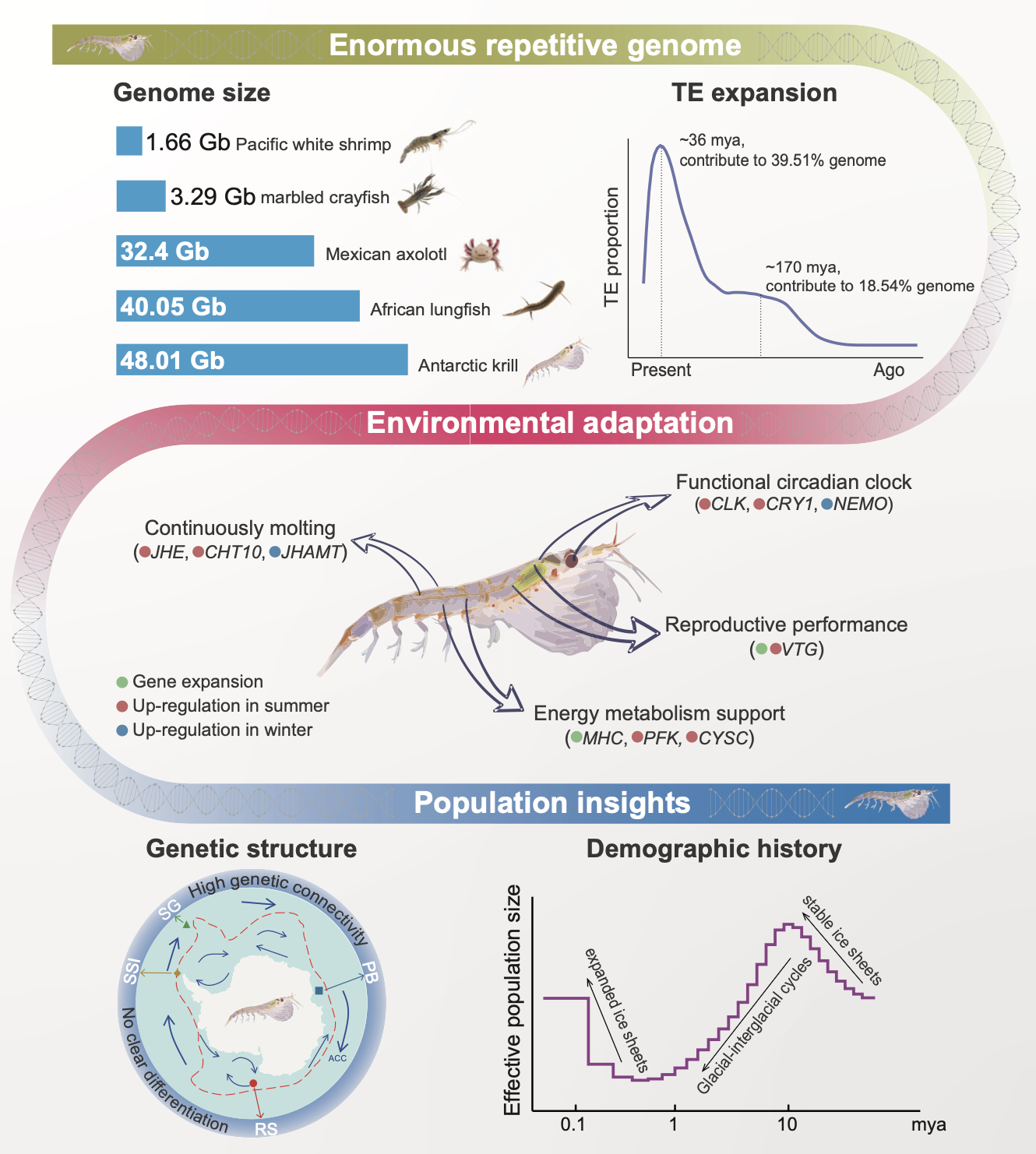
Giant and Repetitive Genome Assembly:
The Antarctic krill genome, dominated by extensive transposable element (TE) expansions, posed significant assembly challenges. Using PacBio long-read sequencing combined with Hi-C data, we achieved a chromosome-level assembly comprising 17 pseudochromosomes and 28,834 annotated protein-coding genes. Repetitive sequences accounted for over 92% of the genome, far exceeding proportions observed in other large genomes, such as the Mexican axolotl or African lungfish. - Environmental Adaptations:
Our analysis uncovered genetic adaptations that support krill’s survival in the extreme Antarctic environment:- Circadian Rhythm Genes: Genes like CLOCK, CRY1, and NEMO exhibit seasonally modulated expression, revealing how krill synchronize life-cycle events with environmental changes.
- Molting and Energy Metabolism Genes: Expanded gene families associated with chitin metabolism, energy regulation, and molting enable krill to thrive despite fluctuating food availability and extreme environmental conditions.
- Population Structure and Dynamics:
Genome-wide resequencing of 75 individuals from four geographic locations revealed no significant population structure, emphasizing high genetic connectivity across the Southern Ocean. However, we identified subtle signals of local adaptation, potentially shaped by environmental selection. Demographic history reconstruction indicates population stability during glacial periods and subsequent expansion over the past 100,000 years.
Reflections and Full-Text Access
This publication involved the analysis of a massive dataset and collaboration with our partners at the Yellow Sea Fisheries Research Institute. Despite challenges in achieving continuity during genome assembly due to the highly repetitive nature of the genome, our use of third-generation long-read sequencing paired with Hi-C data enabled chromosome-level assembly. Genome annotation was equally challenging, particularly in identifying and analyzing repetitive sequences. We compared Antarctic krill genome features with those of other large genomes and focused on annotating circadian rhythm and molting-related genes.
The population resequencing project was especially demanding due to the genome’s size, requiring extensive data processing. Understanding krill population structure was crucial, given their ecological significance. I was deeply involved in the long peer-review process, addressing reviewers’ comments in extensive detail, with each round of revisions spanning dozens of pages. This experience reaffirmed the value of persistence and collaboration in achieving scientific breakthroughs.
The full text of this study is available online at Cell.