mtscATAC-seq based on DNBelab C4 published
Our recent paper in Aging Cell described our recently developed single-cell mitochondrial sequencing methods. Here we provide major information on this paper, as well as the method.
Mitochondria play a crucial role in various biological processes, and their dysfunction is often associated with aging and age-related diseases. However, studying mitochondrial heterogeneity at the single-cell level has been challenging due to limited methods and resources. Our lab has recently made significant strides in this area by developing a novel single-cell mitochondrial sequencing method, C4_mtscATAC-seq, which allows us to investigate mitochondrial DNA (mtDNA) mutations and chromatin accessibility simultaneously.
Research Background
Mitochondria are essential organelles involved in energy production, metabolism, and cell signaling. During the aging process, mitochondrial function declines, leading to a decrease in oxidative phosphorylation activity, accumulation of mtDNA mutations, and increased production of reactive oxygen species. Understanding the dynamics of mitochondrial changes at the single-cell level can provide valuable insights into the aging process and potential therapeutic targets.
Methodology
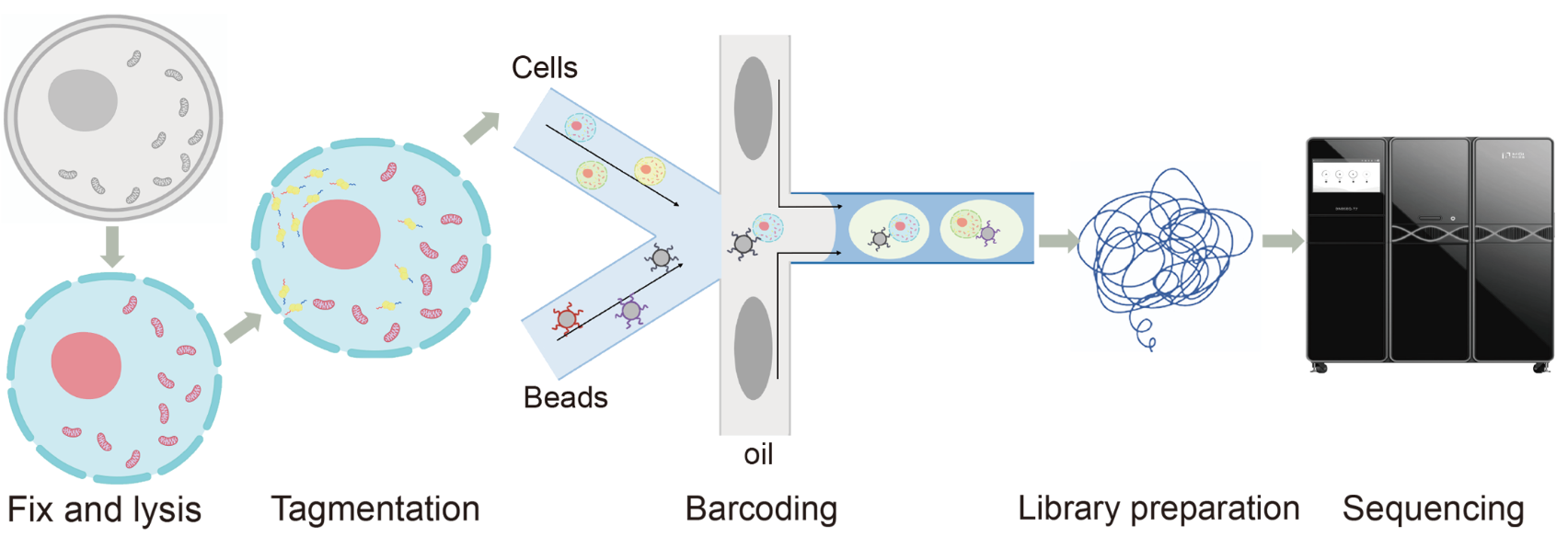
Our lab optimized the DNBelab C4 single-cell ATAC sequencing workflow to develop the C4_mtscATAC-seq protocol. This method enables the simultaneous capture of mitochondrial content and open chromatin status in individual cells. We validated our protocol using the HEK-293T cell line and then applied it to study two mouse tissues: spleen and bone marrow, from both young (2 months old) and aged (23 months old) mice.
Key Findings
Higher mtDNA Content in Young Tissues: We found that young tissues had higher mtDNA content compared to aged tissues, consistent with higher activity scores of nuclear genes associated with mitochondrial replication and transcription in young tissues. Low-Frequency mtDNA Mutations: Our study identified a total of 22, 15, and 21 mtDNA mutations in the young spleen, aged spleen, and bone marrow, respectively. Most of these mutations had variant allele frequencies (VAF) below 1%. Age-Related Increase in mtDNA Mutations: Aged tissues exhibited a higher number of mtDNA mutations with higher VAF compared to young tissues. We identified three mtDNA variations (m.9821A>T, m.15219T>C, and m.15984C>T) with the highest VAF in both aged spleen and bone marrow. Differential Chromatin Accessibility: By comparing cells with and without these mtDNA variations, we analyzed differential open chromatin status to identify potential genes associated with these variations, including transcription factors such as KLF15 and NRF1.
Implications and Future Work
Our study provides an alternative single-cell mitochondrial sequencing method and offers insights into age-related mitochondrial variations. The findings suggest that mitochondrial dysfunction and mtDNA mutations may play a significant role in aging. Future work will focus on further exploring the molecular mechanisms underlying these variations and their impact on cellular function.
Conclusion
The development of C4_mtscATAC-seq represents a significant advancement in the field of mitochondrial genomics. Our lab is committed to continuing research in this area to deepen our understanding of mitochondrial dynamics in aging and other biological processes. We believe that our findings will contribute to the development of new strategies for mitigating age-related diseases and promoting healthy aging.