Massive Genomic Study Catalogues Marine Microbes
🌊 BGI Research Team Unveils Vast Marine Microbial Diversity and Biotechnological Potential in Nature and successfully discovers novel gene editing tools, antibiotics, and plastic-degrading enzymes.
The world’s oceans harbor a staggering diversity of microbial life, a largely untapped resource with immense potential. A major study published online in Nature, involving researchers from BGI Research, Shandong University, University of East Anglia, and other international collaborators, presents a landmark effort to catalogue this diversity and unlock its biotechnological applications. Key BGI Research contributors include Guangyi Fan, Wenwei Zhang, Xin Liu, Ying Sun, Jianwei Chen, and Yangyang Jia.
💡 The Challenge: From Data to Discovery
While sequencing technology has generated vast amounts of marine microbial genomic data, translating this into practical biotechnological or biomedical applications has remained difficult. This study aimed to bridge that gap.
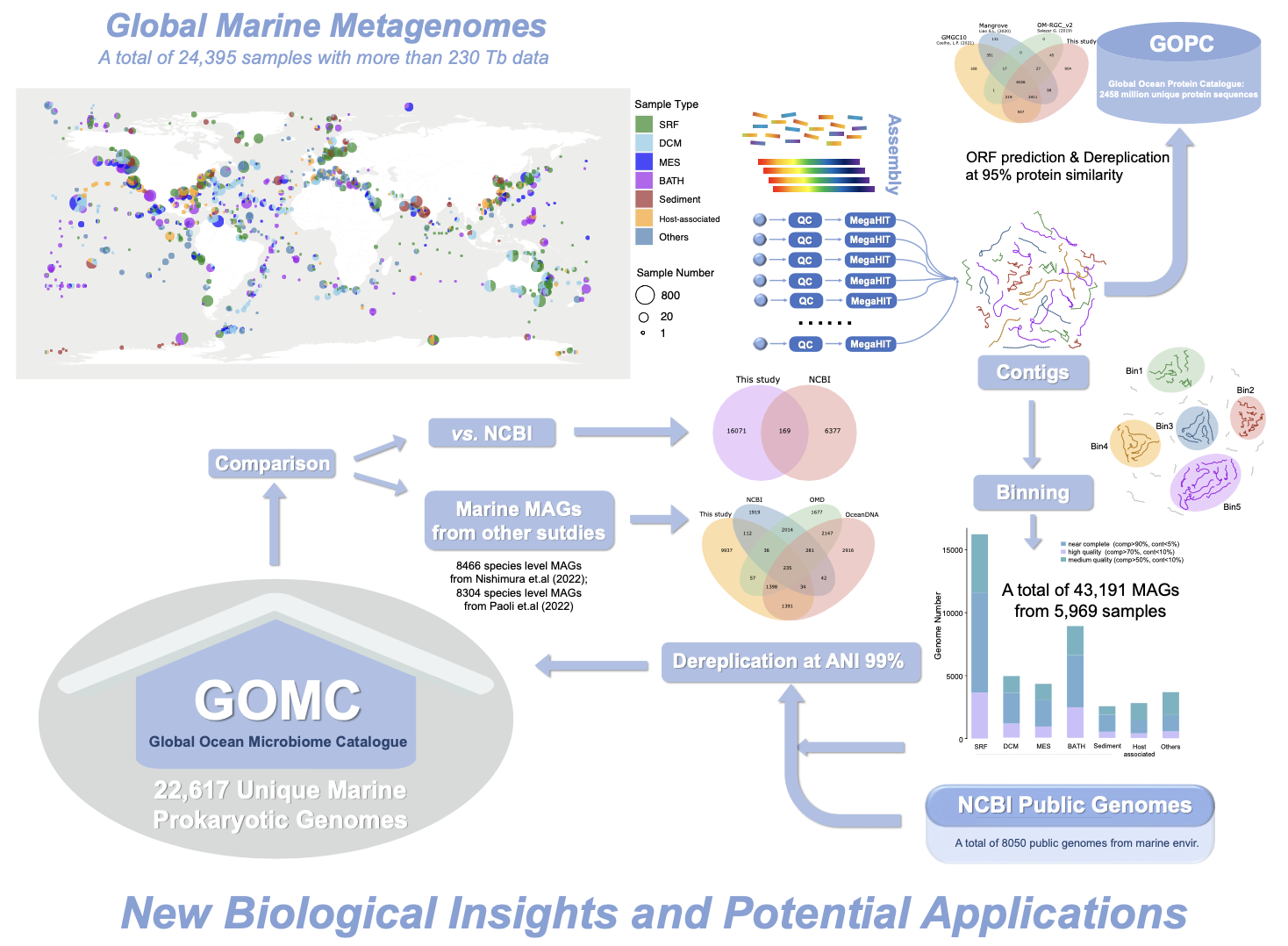
🌍 Building a Global Ocean Microbiome Catalogue (GOMC)
The research team analyzed enormous volumes of public marine metagenome data, resulting in:
- 🧬 Genome Recovery: Reconstruction of 43,191 unique microbial genomes (MAGs) from diverse marine environments.
- 📖 Unified Catalogue (GOMC): Integration with existing databases to create the GOMC, containing nearly 24,200 unique species-level genomes, significantly expanding known marine microbial diversity.
- 🦠 Novel Insights: Characterization of genomic features, including the largest known marine bacterial genomes (up to 18.4 Mb) and complex relationships between microbial defense systems (CRISPR-Cas vs. antibiotic resistance genes).
✨ Bioprospecting Success: Mining Marine Genomes
The true power of this resource was demonstrated through successful in silico bioprospecting combined with rigorous experimental validation:
- ✂️ Novel CRISPR-Cas9 System: Discovered Om1Cas9, a compact and efficient new gene-editing tool, and confirmed its activity in vitro and in human cell lines.
- 💊 New Antimicrobial Peptides (AMPs): Identified over 1,000 potential AMPs. Ten synthesized candidates showed antibacterial activity, including cAMP_87, which exhibited potent, broad-spectrum action against pathogens.
- ♻️ Potent Plastic-Degrading Enzymes: Found novel PETases (enzymes that break down PET plastic) from deep-sea environments. Three candidates displayed superior PET degradation activity, especially under high-salt conditions, outperforming known enzymes.
🚀 Significance: A Resource for the Future
This comprehensive study not only provides an invaluable resource – the GOMC and associated protein catalogue (GOPC) – for fundamental research into marine ecosystems and microbial evolution, but it also powerfully demonstrates that global genomic data can be effectively mined to discover tangible solutions for medicine, biotechnology, and environmental remediation.
The success in identifying and validating novel gene editors, potential antibiotics, and plastic-degrading enzymes highlights the immense, accessible potential residing within the ocean’s microbiome.
💡 Personal thinking
This project originated after the establishment of the Qingdao branch, where there was an urgent need to carry out foundational work in the marine field, particularly related to databases. As a result, we initiated efforts to organize and summarize existing data across various aspects of marine genomics. In 2018, we compiled summaries for plants, animals, and microorganisms, culminating in the creation of a marine white paper. This document not only consolidated publicly available genomic data up to that point but also explored potential future directions in the field of marine genomics.
The primary author of this paper, Chen Jianwei, was responsible for summarizing the microbial content of the white paper, as he was already leading environmental metagenomics research at BGI. Under his leadership, the team comprehensively collected and organized marine microbial data from public databases, creating a complete dataset. They conducted de novo assembly and rederived functional elements of the microorganisms. Subsequent work involved collaboration with partners on data mining and functional validation, which ultimately led to the publication of this paper through joint efforts.
The key to the project’s success lies in leveraging large-scale datasets and maintaining long-term perseverance in tackling challenging tasks. This approach proved essential to achieving these significant outcomes.
📄 Publication Details
- Paper: Chen, J., Jia, Y., Sun, Y. et al. Global marine microbial diversity and its potential in bioprospecting. Nature (2024).
- Published Online: 04 September 2024
- DOI: 🔗 https://doi.org/10.1038/s41586-024-07891-2
- Data & Resources: 🌐 Explore the data interactively at CNGBdb: https://db.cngb.org/maya/datasets/MDB0000002