Multi-omics Landscape and Molecular Basis of Radiation Tolerance in a Tardigrade
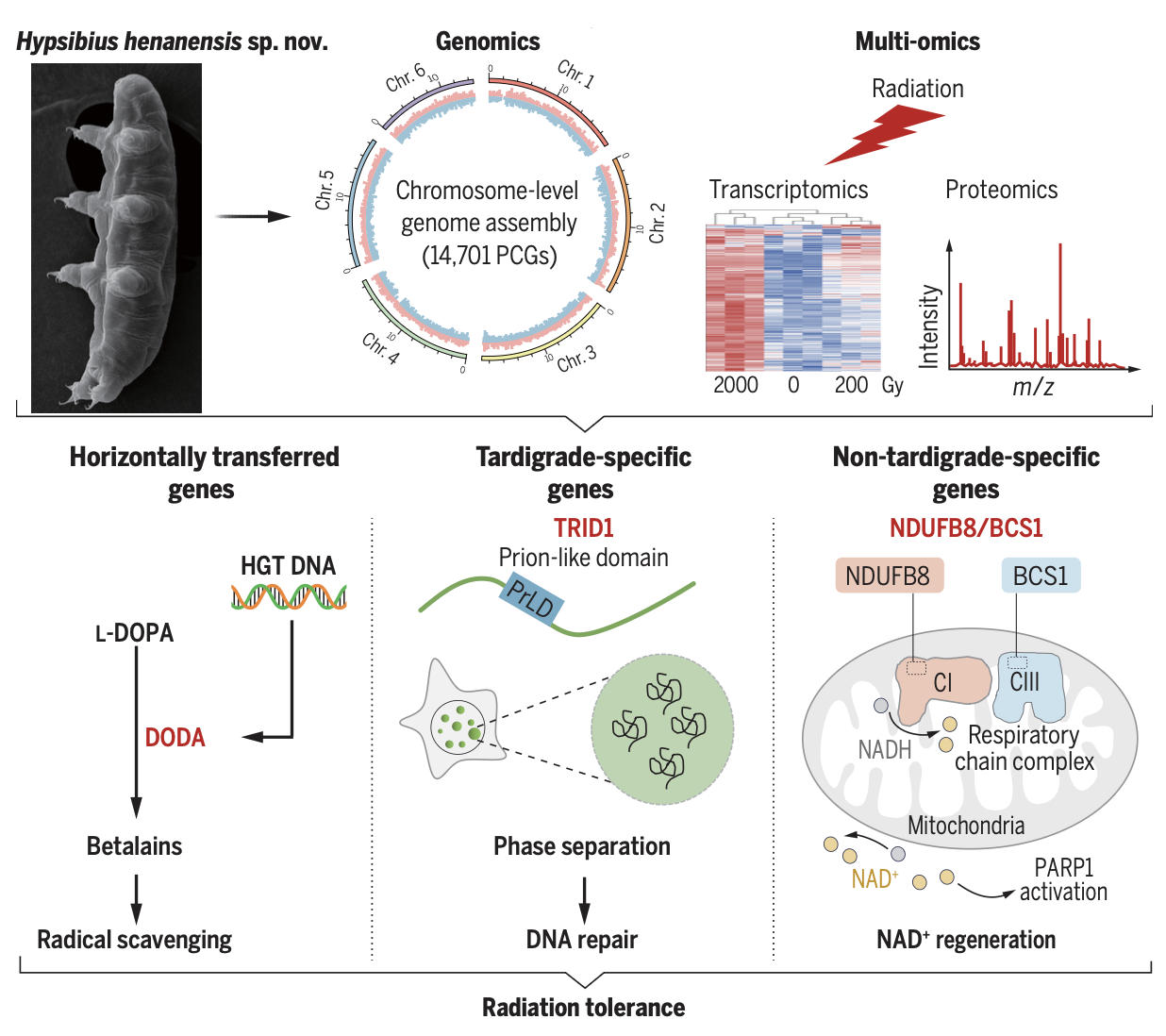
In our latest collaborative project, we delved deep into the molecular basis of radiation tolerance in a newly identified tardigrade species, Hypsibius henanensis sp. nov. Using a comprehensive multi-omics strategy—including genomics, transcriptomics, and proteomics—we sought to unravel the mechanisms that allow tardigrades to thrive under extreme ionizing radiation.
Key Findings
-
Chromosome-Level Genome Assembly and Bioinformatics Analysis:
We generated a high-quality, chromosome-level reference genome for H. henanensis sp. nov. Through rigorous bioinformatics analysis, we identified 14,701 protein-coding genes and cataloged 2801 differentially expressed genes (DEGs) in response to heavy ion radiation. -
Mechanisms of Radiotolerance:
Our data mining efforts uncovered several key mechanisms:- A horizontally transferred gene (DODA1)—encoding a DOPA dioxygenase responsible for betalain biosynthesis—appears to mitigate reactive oxygen species (ROS) through pigment production.
- A tardigrade-specific radiation-induced disordered protein (TRID1) promotes efficient DNA double-strand break (DSB) repair through liquid-liquid phase separation (LLPS), thereby enhancing cell survival under extreme conditions.
- Additionally, non-tardigrade-specific genes, such as mitochondrial proteins involved in NAD⁺ regeneration (e.g., BCS1 and NDUFB8), further contribute to the radioprotective response.
Our Collaborative Role and Personal Impressions
In this project, XinLab primarily contributed the robust bioinformatics analysis that helped characterize the multi-omics landscape, complementing the extensive experimental validations performed by our collaborators. I still remember the excitement upon first reading the initial manuscript draft—even though there were many suggested revisions from my side, the experimental team maintained the core message that impressed the Science editors. The review process was exceptionally smooth and well-coordinated, making it one of the most efficient encounters I’ve experienced.
Although XinLab’s role was supportive rather than central (with Guangyi Fan serving as the co-corresponding author), I find it very gratifying to be part of such a compelling genomics study. A particular highlight was seeing our names—along with our Chinese names—listed on the final Science article. This detail may seem small, but it was a source of genuine pride for me.
Conclusion
Our multi-omics study not only broadens our understanding of how tardigrades survive extreme radiation but also underscores the versatility of bioinformatics in uncovering complex stress tolerance mechanisms. We hope that these insights will inspire further research into cell stress responses and ultimately contribute to advancements in biomedical and environmental sciences.