Resequencing 50 Accessions of Cultivated and Wild Rice for Agronomic Gene Identification
Rice (Oryza sativa) is a staple food crop that has undergone extensive domestication, resulting in significant phenotypic and physiological changes. Understanding the genetic variation between cultivated and wild rice is key to identifying valuable agronomic traits, optimizing breeding strategies, and improving crop yield and stress resilience. In our latest study, published in Nature Biotechnology, we resequenced 50 rice accessions, covering 40 cultivated and 10 wild varieties, generating a high-density single-nucleotide polymorphism (SNP) dataset to uncover candidate genes selected during domestication.
Key Findings
- Genome-Wide Variation Analysis:
- Resequencing efforts yielded 6.5 million high-quality SNPs, providing the most comprehensive variation dataset for rice genome research.
- Comparative analysis between cultivated and wild rice identified genomic regions with significantly reduced diversity, suggesting human-driven selection events.
- Domestication and Population Structure:
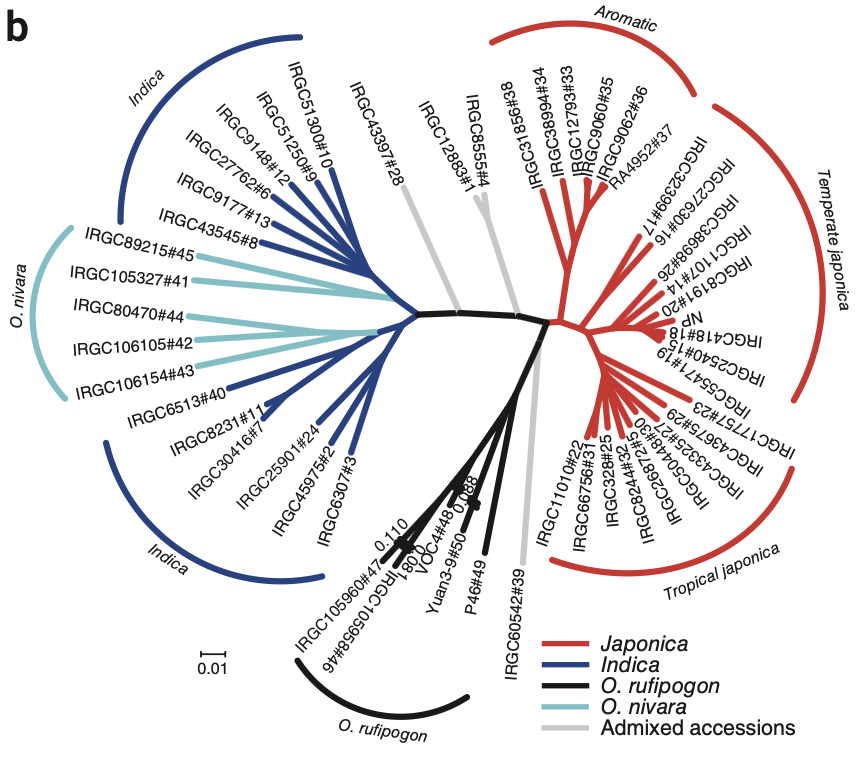
- Phylogenetic analyses confirmed independent domestication pathways for indica and japonica rice, aligning with wild ancestors O. nivara and O. rufipogon, respectively.
- Principal component analysis (PCA) and neighbor-joining trees highlighted clear genetic differentiation between rice subspecies, reinforcing distinct evolutionary trajectories.
- Candidate Genes Under Selection:
- We detected 739 selective sweep regions in japonica and 750 in indica rice, pinpointing genes related to plant architecture, flowering, and stress tolerance.
- Key domestication genes such as prog1 (growth regulation) and sh4 (seed shattering suppression) were successfully identified.
- Applications for Rice Breeding and Genetic Improvement:
- Genome-wide SNP markers serve as an essential tool for marker-assisted selection, enabling breeders to develop high-yield, stress-resistant rice varieties.
- Functional annotation of candidate genes highlights potential targets for improving grain quality, disease resistance, and abiotic stress tolerance.
Reflections
When I took over this project, it had already undergone an initial round of peer review at Nature and was undergoing major revisions. These included increasing sequencing depth and expanding sample selection with additional individuals for re-sequencing. Ultimately, we analyzed 50 rice varieties, alongside 15 additional varieties sequenced at lower depths, making this study one of the larger-scale population genomic analyses in rice research. Despite the challenges of revision and refinement, the final publication in Nature Biotechnology represents a significant contribution to rice genomics and crop improvement.
The full text of this study can be accessed online at Nature Biotechnology.