Nature Genetics: Maize HapMap2 Unlocks the Extensive Variation in a Genome in Flux
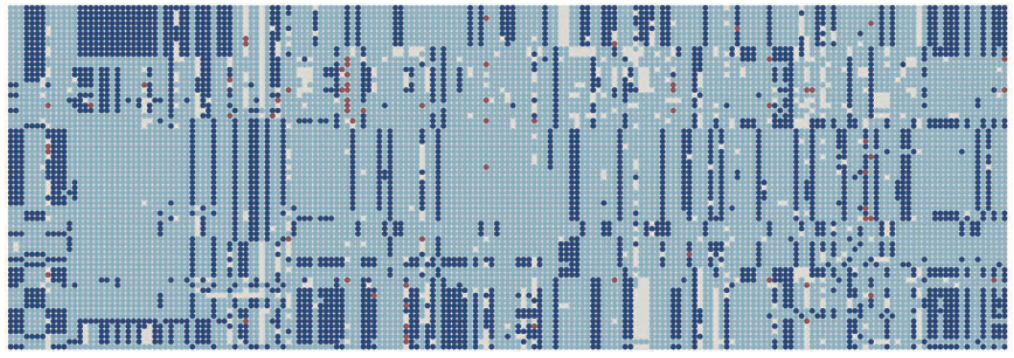
Our 2012 study in Nature Genetics characterized 55 million SNPs across 103 maize varieties, providing a transformative resource to identify beneficial alleles from undomesticated varieties.
The study “Maize HapMap2 identifies extant variation from a genome in flux,” co-authored by researchers including Xin Liu (XinLab), was published in Nature Genetics.
Background
Maize (Zea mays) is a cornerstone of global agriculture, but its genome is notoriously complex, characterized by high levels of repetitive sequences and structural variations. While modern breeding has significantly improved yields, progress in tapping into the genetic diversity of wild teosinte and landraces remained limited due to the lack of high-density genetic markers.
Key Breakthroughs
- 55 Million SNPs: Utilizing a population genetics scoring model, we identified 55 million SNPs in a diverse panel of 103 lines. This remains one of the most comprehensive catalogs of genetic variation for any crop species.
-
A Genome in Flux: The study highlighted the dynamic nature of the maize genome, where transposable elements and frequent rearrangements drive extensive variation between individuals.
- Domestication and Selection Signatures: By comparing domesticates with their wild relatives, we mapped specific genomic regions subjected to intense artificial selection. Our findings underscored the value of wild teosinte as a reservoir of beneficial alleles for future crop resilience.
- Structural Variation Profiling: Beyond simple SNPs, the project analyzed insertions, deletions, and copy number variations (CNVs), revealing their profound impact on gene expression and complex agronomic traits.
Impact & Significance
The Maize HapMap2 provides the essential foundation for next-generation precision breeding. By enabling the identification of elite alleles from undomesticated varieties, it facilitates the development of corn varieties with improved nutritional profiles and enhanced tolerance to environmental stress.
Collaborative Effort
This milestone project was a massive international collaboration involving Cold Spring Harbor Laboratory (CSHL), Cornell University, BGI, and several other leading institutions worldwide.
Read the full article here: https://doi.org/10.1038/ng.2313