Genomic Variation Map of Cucumber: Insights into Domestication and Diversity
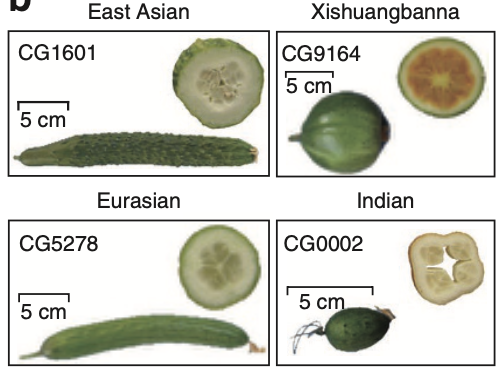
Most fruits consumed today are the result of long-term domestication and breeding efforts. However, the genetic basis underlying domestication traits and population diversity remains largely unexplored in several fruit crops. In our latest study, published in Nature Genetics, we present a genome-wide variation map of cucumber, generated through deep resequencing of 115 cucumber lines sampled from 3,342 accessions worldwide. Our findings offer new perspectives on cucumber domestication, trait selection, and genomic diversity, providing valuable insights for future breeding programs.
Key Findings
- Comprehensive Genome-Wide Variation Analysis:
- Deep resequencing of 115 cucumber lines revealed ~3.6 million genetic variants, including 3,305,010 SNPs, 336,081 small indels, and 594 presence-absence variations (PAVs).
- Comparative analysis with the wild cucumber genome (Cucumis sativus var. hardwickii) identified 21,021 orthologous genes, helping to refine the evolutionary history of cucumber.
- Domestication Bottlenecks in Fruit vs. Grain Crops:
- Cucumber underwent a much narrower population bottleneck during domestication compared to grain crops like rice and maize, leading to a greater loss of genetic diversity.
- The estimated effective population size at domestication was ~500, which is significantly lower than that of maize (~150,000) and rice (~1,300).
- Identification of Domestication Sweeps:
- A total of 112 domestication sweep regions were detected, spanning 7.8% of the genome (15.4 Mb) and affecting 2,054 genes.
- Notably, one of these regions contains a gene associated with the loss of fruit bitterness, a critical trait selected during cucumber domestication.
- Natural Genetic Variation for Nutritional Improvement:
- The study identified a natural genetic variant in a β-carotene hydroxylase gene (CsaBCH1) responsible for β-carotene accumulation in Xishuangbanna cucumbers.
- This discovery opens new possibilities for breeding cucumbers with enhanced nutritional value, particularly varieties rich in provitamin A.
Reflections
Collaborating with Professor Sanwen Huang on this project was an incredibly rewarding experience, as I gained a deeper understanding of cucumber history and cultivation practices. Additionally, I learned how to efficiently drive large-scale genomics projects forward, which proved invaluable for future research. Alongside Peng Zeng, I was responsible for data analysis, and the insights we uncovered significantly advanced our knowledge of cucumber domestication and genetic diversity.
The full text of this study can be accessed online at Nature Genetics.