Pearl Millet Genome Sequencing: A Resource for Improving Agronomic Traits in Arid Regions
Pearl millet (Cenchrus americanus) is a staple crop for over 90 million farmers in arid and semi-arid regions, particularly in sub-Saharan Africa and South Asia. Its resilience to extreme environmental conditions, such as high temperatures, low soil moisture, and limited rainfall, makes it a crucial crop for ensuring food security in a warming climate. Despite its agricultural significance, genomic resources for pearl millet have remained limited—until now.
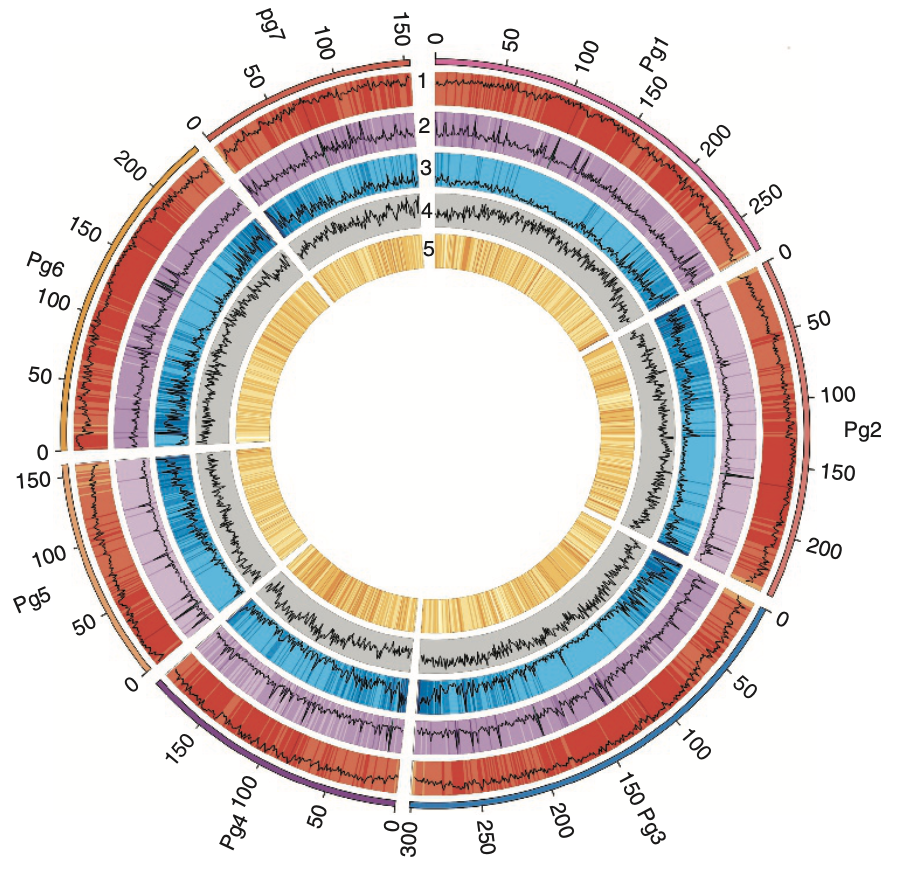
In our latest study, published in Nature Biotechnology, we assembled a 1.79 Gb draft genome for the reference genotype Tift 23D2B1-P1-P5, identifying 38,579 protein-coding genes and uncovering key genetic adaptations that contribute to heat and drought tolerance. Our large-scale resequencing of 994 pearl millet lines provided deep insights into population diversity, domestication patterns, and hybrid performance prediction, offering a valuable genomic foundation for breeding programs aimed at improving grain yield, stress resilience, and agronomic traits.
Key Findings
- Genome Assembly and Annotation:
- Achieved high-quality genome assembly covering 90% of the estimated 1.76 Gb genome.
- Identified gene families linked to lipid biosynthesis, contributing to drought and heat tolerance.
- Detected 38,579 protein-coding genes, with key expansions in cuticular wax biosynthesis pathways, vital for survival in high-temperature environments.
- Genetic Diversity and Domestication Insights:
- Resequencing 994 lines, including 31 wild accessions, revealed strong population structure among wild genotypes, tracing domestication origins to central West Africa.
- Identified 140 genomic regions under selection pressure, including those linked to inflorescence morphology and growth traits, demonstrating genetic adaptation throughout domestication.
- Marker-Trait Associations and Genomic Selection:
- Conducted genome-wide association studies (GWAS) for 20 key agronomic traits, pinpointing 1,054 significant genetic markers for grain yield, plant height, panicle size, and stress adaptation.
- Genomic selection models successfully predicted high-yield hybrid combinations, enabling breeders to accelerate selection cycles for improved hybrids.
Reflections
This project took us through an arduous publication journey. After spending more than a year under review at Nature, we faced rejection—an incredibly painful and frustrating experience. However, perseverance led us to the eventual successful publication in Nature Biotechnology, which remains a strong and well-respected journal in the field. Despite the setbacks, the impact and recognition of this work make the challenges worthwhile.
The full text of this study can be accessed online at Nature Biotechnology.