Draft Genomes of Five African Orphan Crops: Expanding Genetic Resources for Food Security
To diversify global food supplies and enhance nutrition security, underutilized crops—often referred to as orphan crops—offer an essential path forward. Unlike major staple crops such as wheat, rice, and maize, these species remain largely overlooked in modern breeding programs despite their economic, nutritional, and environmental significance. In our latest study, published in GigaScience, we present draft genome assemblies for five African orphan crops, shedding light on their genetic diversity and potential for agricultural improvement.
Key Findings
- Genome Sequencing and Assembly of Five Species:
We successfully sequenced and assembled the genomes of:- Vigna subterranea (Bambara groundnut): A drought-tolerant legume rich in protein and carbohydrates.
- Lablab purpureus (Hyacinth bean): A multipurpose legume with nitrogen-fixing abilities.
- Faidherbia albida (Apple-ring acacia): A nitrogen-fixing agroforestry tree adapted to arid regions.
- Sclerocarya birrea (Marula): A fruit-bearing tree with nutritional and medicinal applications.
- Moringa oleifera (Moringa): A fast-growing tree known for its leaves’ high protein and antioxidant content.
- Gene Annotation and Functional Insights:
The gene annotations provided high-resolution insights into starch biosynthesis, transcription factor evolution, and nitrogen fixation pathways.- Vigna subterranea exhibited expanded starch metabolism genes, potentially contributing to its high resistant starch content.
- Faidherbia albida showed unique adaptations for root nodule symbiosis, facilitating its nitrogen-fixing capability.
- Comparative genomic analysis uncovered gene families involved in stress resistance, fruit development, and metabolic regulation, providing valuable targets for breeding improvements.
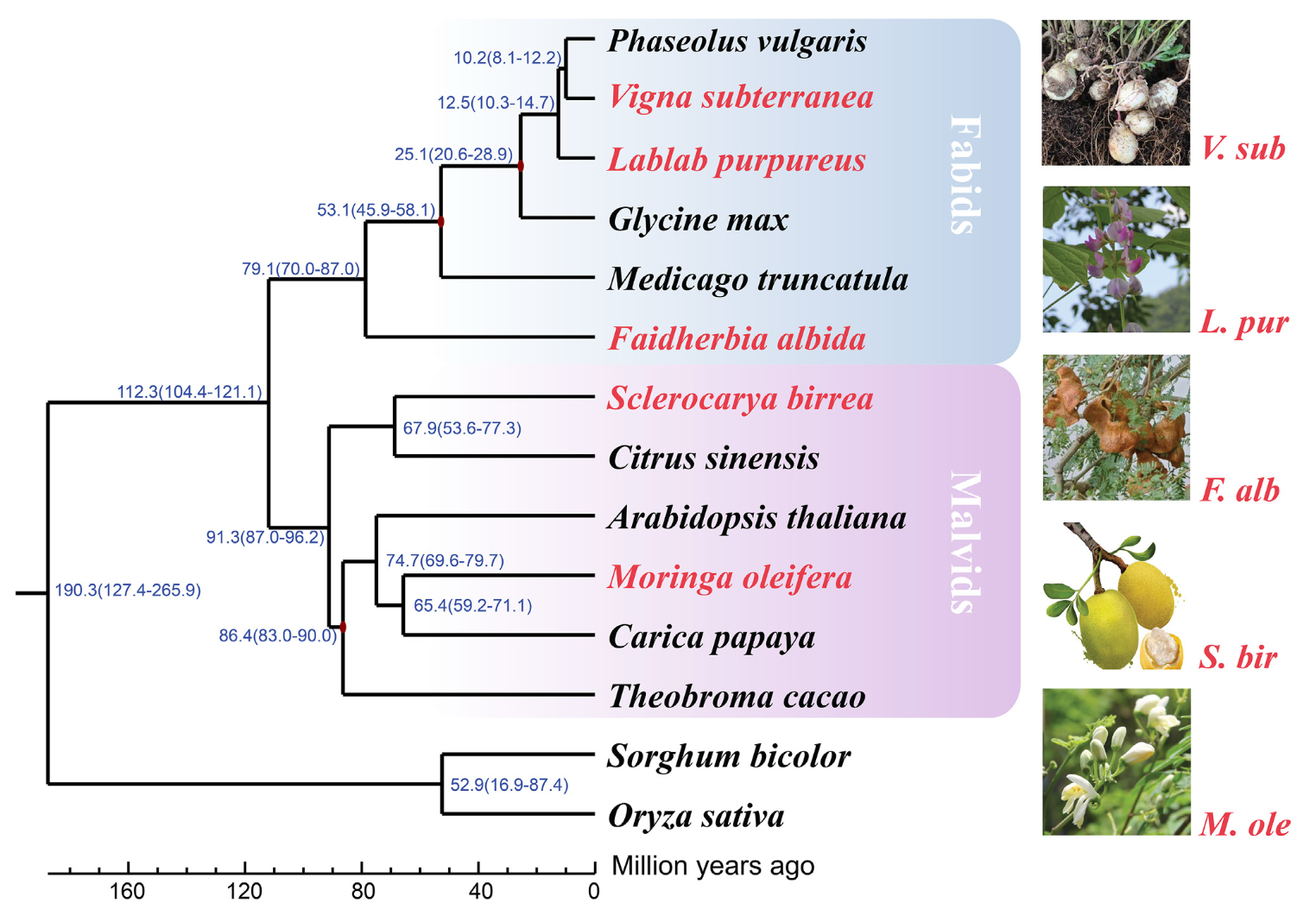
- Phylogenetic Analysis and Evolutionary Divergence:
Using single-copy genes, we constructed a phylogenetic tree, estimating the evolutionary divergence times between these orphan crops and related species.- Moringa oleifera diverged from Carica papaya approximately 65 million years ago, while Faidherbia albida separated from other legumes 79 million years ago.
- Gene family expansion and contraction analyses revealed adaptive genomic features linked to environmental resilience.
Reflections
This research is part of the African Orphan Crop Genome Initiative, which we launched in 2015, aiming to systematically sequence and analyze orphan crops that play crucial roles in African agriculture and food security. The project was conducted in collaboration with UC Davis and the African Orphan Crop Consortium (AOCC), with sample contributions from ICRAF (World Agroforestry Centre). By leveraging high-throughput sequencing technologies, we successfully assembled genome references for five key crops, contributing essential genetic insights for future breeding, conservation, and agricultural development.
Moving forward, our efforts will continue to expand genomic resources for underrepresented crops, supporting sustainable agriculture and improving food security through genomics-driven plant breeding.
The full text of this study can be accessed online at GigaScience.