Resequencing 429 Chickpea Accessions: Insights into Genome Diversity and Domestication
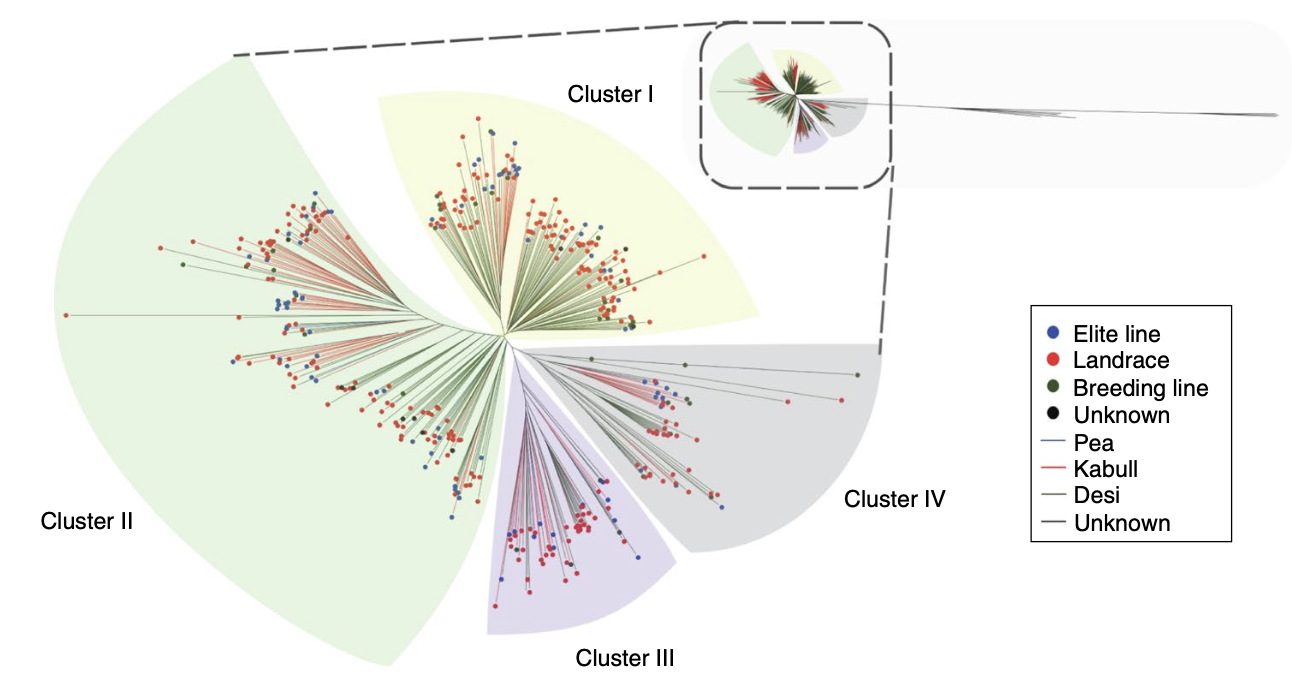
Chickpea (Cicer arietinum) is a staple legume crop with immense nutritional and agricultural importance. Understanding its genetic diversity is crucial for crop improvement, especially in the face of climate challenges. In our latest study, published in Nature Genetics, we conducted whole-genome resequencing of 429 chickpea accessions from 45 countries, uncovering valuable insights into domestication, population structure, and agronomic trait associations.
Key Findings
-
Genome-Wide Variations and Genetic Diversity:
Through resequencing, we identified 4.97 million SNPs, 596,100 small insertions/deletions (indels), and other structural variations across 429 chickpea lines. The data revealed that wild chickpea species exhibit significantly higher genetic diversity than cultivated varieties, with breeding processes narrowing the gene pool. - Domestication and Selection Events:
- Our findings suggest that chickpea originated in the Eastern Mediterranean, with migration routes extending to South Asia, Central Asia, and Ethiopia.
- A strong bottleneck was detected during domestication, with 80% of genetic diversity lost from wild genotypes to landraces.
- We identified 122 genomic regions and 204 genes under selection, many related to stress resistance, flowering time, and seed development—traits shaped by human-driven breeding.
- Genome-Wide Association Study (GWAS) for Agronomic Traits:
- GWAS pinpointed 262 molecular markers and candidate genes for drought and heat tolerance, crucial for breeding climate-resilient chickpea varieties.
- Specific genes linked to early flowering, root architecture, and stress adaptation were discovered, providing targets for marker-assisted breeding strategies.
Reflections
This study is another major achievement in our collaborative research on chickpea genomics with Rajeev Varshney’s team, following the publication of the reference genome. Since the initial genome sequencing, we have been actively conducting resequencing studies to expand our understanding of chickpea’s genetic diversity, adaptation mechanisms, and breeding potential.
Our findings offer crucial genomic resources for breeders, helping accelerate the development of chickpea varieties that are more resilient to environmental stresses and more productive in diverse agro-climatic regions. The discovery of sex-related genomic regions and stress-adaptation genes further highlights the importance of population genomics for applied breeding. While our ongoing work delves deeper into sex-determination mechanisms, this study marks a significant step forward in enhancing chickpea’s genetic foundation for future agricultural sustainability.
The full text of this study can be accessed online at Nature Genetics.