Draft Genomes of Jackfruit and Breadfruit Provide Insights into Starch and Sugar Metabolism
Jackfruit (Artocarpus heterophyllus) and breadfruit (Artocarpus altilis) are two economically and nutritionally significant species within the Artocarpus genus. As staple food crops in tropical regions, their genomic characteristics play a crucial role in understanding their high starch and sugar content, as well as potential applications in food security and crop improvement. In our latest study, published in Genes, we present high-quality draft genome assemblies of jackfruit and breadfruit, offering new insights into their metabolism and evolutionary adaptations.
Key Findings
- Genome Assembly and Annotation:
- A total of 273 Gb and 227 Gb of sequencing data was generated for jackfruit and breadfruit, respectively.
- The jackfruit genome was assembled into 162,440 scaffolds totaling 982 Mb, while the breadfruit genome comprised 180,971 scaffolds totaling 833 Mb.
- Gene annotation identified 35,858 protein-coding genes in jackfruit and 34,010 in breadfruit, including pathways associated with starch and sucrose metabolism.
- Expansion of Starch and Sugar Metabolism Genes:
- Comparative analysis revealed expansion of gene families related to starch biosynthesis and sugar metabolism, highlighting their distinct fruit composition.
- Tissue-specific expression analysis showed variation in UGD1 and starch biosynthesis genes, possibly explaining differences in starch accumulation.
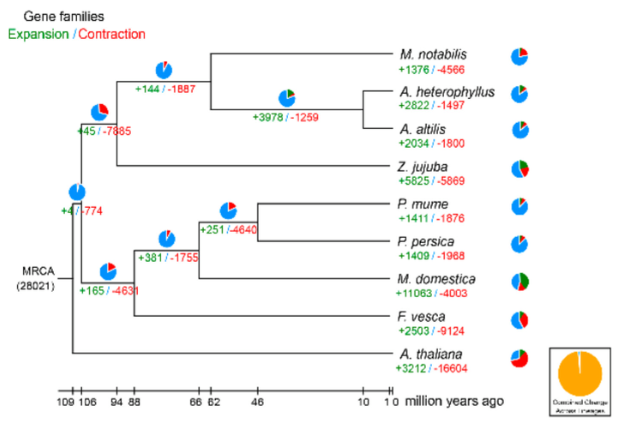
- Evolutionary Insights from Artocarpus Genomes:
- Phylogenetic analysis confirmed that Artocarpus heterophyllus and Artocarpus altilis are closely related to mulberry (Morus notabilis) rather than jujube (Ziziphus jujuba).
- Whole-genome duplication (WGD) analysis suggested that a shared genome duplication event occurred between 62 and 10 million years ago in both species.
Reflections
This research is part of our collaboration with UC Davis and the African Orphan Crop Consortium (AOCC), conducted with sample contributions from ICRAF. The genome sequencing and analysis were led by Bo Song, who efficiently coordinated the project and drove its completion.
As part of broader efforts to sequence underrepresented crops, our team aimed to uncover the genetic basis of starch and sugar metabolism, providing valuable insights for plant breeders and agricultural researchers. Additionally, beyond this study, we are further exploring sex-determination mechanisms in Artocarpus species, which remain a fascinating subject in evolutionary plant biology.
The full text of this study can be accessed online at Genes.