Chromosome-Based Rubber Tree Genome Provides Insights into Evolution and Rubber Biosynthesis
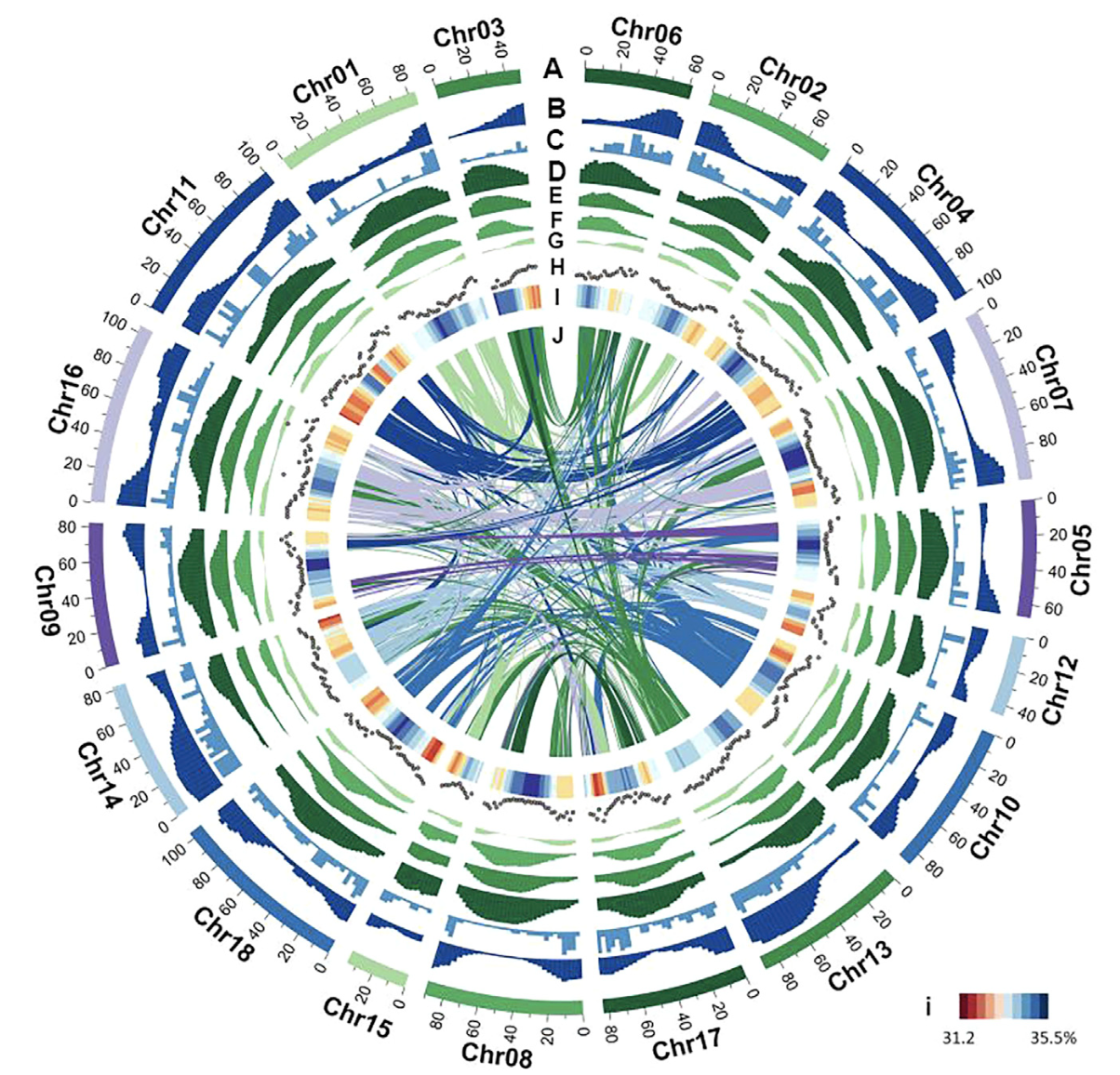
The rubber tree (Hevea brasiliensis) is the primary source of natural rubber, a crucial industrial material with no synthetic equivalent. Understanding the genetic mechanisms underlying rubber production and plant evolution is essential for breeding resilient and high-yielding cultivars. In our latest study, published in Molecular Plant, we present a chromosome-level genome assembly of the rubber tree cultivar GT1, providing key insights into spurge genome evolution, rubber biosynthesis, and domestication history.
Key Findings
-
High-Quality Genome Assembly:
Using PacBio long-read sequencing and Hi-C scaffolding, we generated a 1.47 Gb genome assembly anchored to 18 pseudochromosomes, offering the most complete genomic reference for rubber tree to date. -
Evolutionary Insights into Spurge Genome Structure:
Comparative genomic analysis confirmed a paleopolyploid event predating the divergence of Hevea and Manihot, providing a model for chromosome evolution in the spurge family (Euphorbiaceae). -
Genomic Expansion and Rubber Biosynthesis:
We identified three Hevea-specific LTR retrotransposon families, responsible for genome expansion (~65.88% of the genome). Additionally, large-scale gene expansions were observed in basal metabolic pathways, ethylene biosynthesis, and polysaccharide activation, all essential for latex production. -
Domestication and Genetic Variation:
Whole-genome resequencing of cultivated and wild rubber trees uncovered ~15.7 million SNPs, pinpointing hundreds of candidate domestication genes linked to rubber biosynthesis and agronomic traits.
Reflections
This project is the result of our collaboration with Professor Li-Zhi Gao, focusing on the genome sequencing and genetic study of rubber tree, an economically vital crop. Given the importance of natural rubber, the reference genome and associated datasets presented in this study have garnered substantial attention from researchers and breeders alike.
Our work reinforces the value of chromosome-scale genomic data in unraveling evolutionary relationships and improving crop breeding strategies. By integrating sequencing technologies, comparative genomics, and population studies, we have laid a foundation for future rubber tree research, especially in the areas of biotic and abiotic stress resistance and enhanced rubber production. As we move forward, further studies will build on this genomic resource to deepen our understanding of plant adaptation and molecular mechanisms behind rubber biosynthesis.
The full text of this study can be accessed online at Molecular Plant.