Global Lettuce Genomics: Unraveling the Domestication History and Genetic Diversity of Cultivated Lettuce
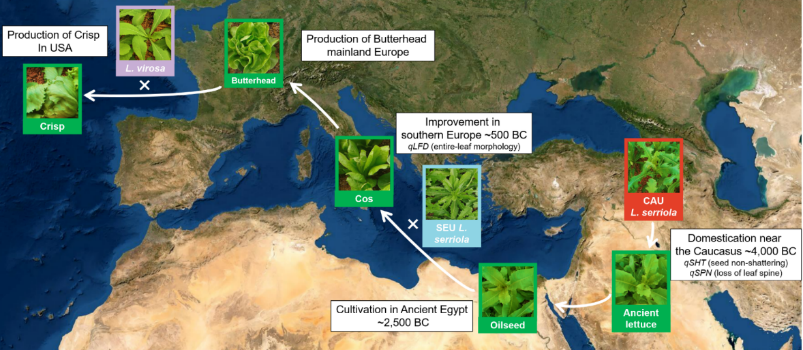
Lettuce (Lactuca sativa) is one of the most widely consumed leafy vegetables in the world, yet its genetic diversity and domestication history have remained largely unexplored. In our recent study, published in Nature Genetics, we performed whole-genome resequencing of 445 Lactuca accessions collected globally, providing a comprehensive genetic variation map for lettuce. These findings offer valuable insights into lettuce evolution, domestication, and potential applications in crop breeding.
Key Findings
-
Reconstructing the Domestication History of Lettuce:
Using phylogenetic and demographic analyses, we traced the origins of cultivated lettuce to the Caucasus region, where initial domestication was marked by the loss of seed shattering. Subsequent cultivation efforts in Southern Europe refined leaf morphology, leading to modern lettuce varieties. -
Genetic Diversity and Structural Variations:
Our sequencing efforts identified 179 million SNPs, 30 million insertions/deletions (indels), and 244,866 structural variants across wild and cultivated lettuce species. This data provides an unprecedented look into how genetic differentiation shaped modern lettuce traits. -
Resistance Clusters and Wild Introgression:
Analysis of major resistance clusters (MRCs) within the lettuce genome revealed extensive wild introgression, specifically from L. serriola, which contributed disease-resistance genes used in breeding for downy mildew resistance. -
Implications for Lettuce Breeding and Crop Improvement:
With a detailed genetic blueprint, this study offers breeders a powerful resource for improving lettuce varieties, enabling enhanced disease resistance, stress tolerance, and yield optimization.
Reflections
After establishing collaborations with our Dutch partners, we worked to advance both genomic sequencing and phenotypic characterization of lettuce. Our collaborators had already collected a global diversity panel of lettuce varieties, and after thorough discussions, we selected representative samples for sequencing. Addressing logistics, we successfully transported the samples to BGI-Shenzhen for genome sequencing.
Additionally, we recognized the importance of conducting field trials outside of Europe to examine environmental effects on plant traits. Leveraging BGI’s agricultural base in Laos, we initiated lettuce germplasm cultivation, collecting phenotypic data over two years—ultimately driving this publication to completion.
This study represents only the beginning of our research into global lettuce populations. A larger-scale sequencing project, involving nearly all known lettuce varieties, is already in progress. Wei Tong, from BGI Research, played a pivotal role in managing international collaborations, sample coordination, data analysis, and manuscript development—helping bring this project to fruition.
The full text of this study can be accessed online at Nature Genetics.