A Comprehensive Chickpea Genetic Variation Map: Insights from 3,366 Genomes
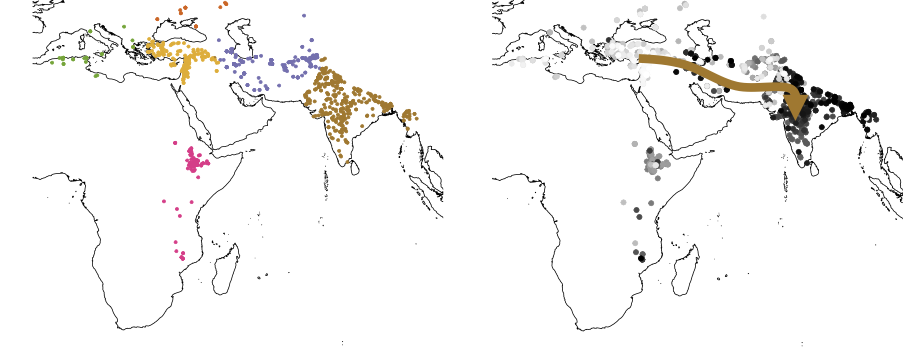
Chickpea (Cicer arietinum), one of the world’s most important legume crops, has long been a focal point in agricultural genomics. In our latest collaborative study with Professor Rajeev Varshney’s team, we conducted whole-genome sequencing on 3,366 chickpea accessions, constructing the most comprehensive chickpea genetic variation map to date. This landmark study provides crucial insights into genetic diversity, enhances trait mapping, and accelerates breeding efforts for climate-resilient chickpea varieties.
Key Findings
-
Extensive Genome Sequencing and Diversity Analysis:
Using high-depth sequencing data, we characterized the genetic diversity across global chickpea populations, identifying millions of SNPs and structural variants that differentiate landraces, wild species, and elite cultivars. -
Accelerating Breeding Through Functional Genomics:
Leveraging genomic variation data, we identified key genetic loci associated with important agronomic traits, including drought tolerance, flowering time, and seed size. These findings provide new opportunities for marker-assisted selection and targeted gene editing in chickpea improvement programs. -
Long-Term Field Trials and Phenotypic Data Integration:
The project combined large-scale genomic data processing with extensive field experiments conducted by our collaborators. By integrating genotypic and phenotypic datasets, we enhanced trait association precision, reinforcing genomic selection strategies for breeding.
Reflections
This publication marks yet another milestone in XinLab’s ongoing research on chickpea genomics. Our collaboration with Rajeev Varshney’s team has been central to advancing genomic resources for legume crops. The project itself spanned a significant timeframe—not only due to extensive genomic sequencing and data analysis but also because of the large-scale field trials and phenotypic data collection undertaken by our collaborators.
Interestingly, this was not our first attempt at publishing a large-scale crop genomic study in Nature. Our previous collaboration with Rajeev’s team involved a pearl millet genome study, which underwent three rounds of peer review over a year before the editor ultimately declined the submission (the study was later published in Nature Biotechnology in 2017). This time, however, we were fortunate—after several rounds of revisions, our chickpea paper was successfully accepted. As we celebrate this achievement, we are already advancing new whole-genome sequencing projects on additional chickpea varieties, with further analyses well underway.
The full text of this study can be accessed online at Nature.