Dynamic TE Shaping Plant Evolution and Crop Traits
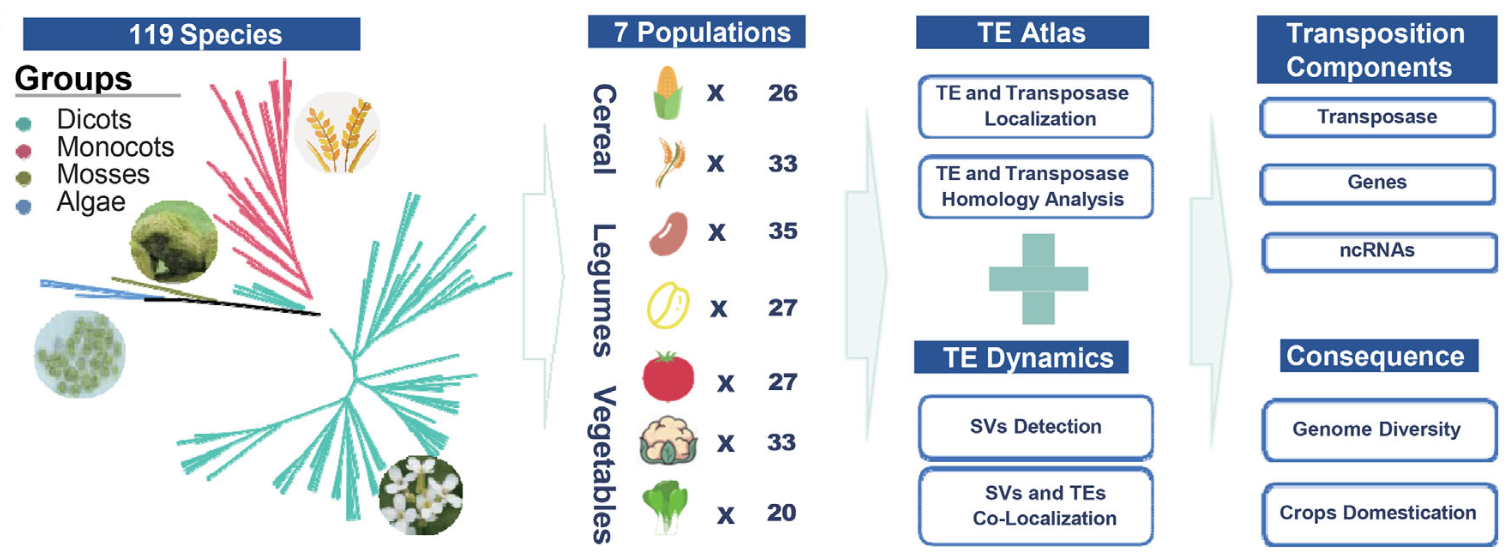
We analyzed 811 plant genomes across 119 species to create an unprecedented atlas of recent transposable element activity, offering insights into genome evolution and crop domestication.
Transposable elements (TEs), often called “jumping genes,” are DNA sequences that can move within a genome. They are major drivers of genetic change, but understanding their recent activity and impact across the plant kingdom has been challenging. A new study published in the Plant Biotechnology Journal by researchers including Xin Liu and Sunil Kumar Sahu from BGI Research tackles this challenge head-on.
By comparing a massive dataset of 811 high-quality plant genome assemblies – spanning 119 diverse species from algae to crops like rice, maize, and tomato – the team has deciphered recent patterns of TE movement. This large-scale comparative genomics approach, enabled by advancements in sequencing and data sharing, provides a detailed look into how these mobile elements have shaped plant genomes.
Key Findings:
- A Vast Atlas of TE Activity: The researchers performed new TE annotations and identified over 13.8 million structural variants in genomes caused by recent TE activity (TE-SVs). This allowed them to map the activity of different TE types (like Copia, Gypsy, Helitron, etc.) across species and within specific crop populations, revealing distinct patterns. For example, Class I retrotransposons were more active in maize and soybean, while Helitron DNA transposons were highly active in cabbage and mustard.
- TEs and Host Genomes Interact: The study provides evidence for a co-evolutionary relationship between TEs and their host genomes. It found that host genes and non-coding RNAs (ncRNAs) are often located near active TEs and are involved in processes like chromatin regulation, DNA binding, and defense responses, suggesting they may influence or be influenced by TE transposition.
- TEs Drive Genetic Innovation: The analysis showed that TEs are a major source of genetic variation, responsible for a significant portion (~60-70%) of structural differences between genomes in the studied crop populations. Active TEs were found to promote the duplication of host genes and frequently insert into regulatory regions, potentially altering gene expression.
- Impact on Crop Domestication & Adaptation: Genes influenced by recent TE activity were found to be linked to important plant functions, including organ development, nutrient absorption, storage metabolism, and responses to environmental stress. This highlights the role TEs have likely played in the domestication of crops and their adaptation to diverse environments.
Significance:
This comprehensive TE dynamics atlas serves as a valuable resource for the plant science community. It illuminates the intricate interplay between mobile elements and host genomes, revealing how TEs contribute to genetic diversity and adaptation. The findings offer a foundation for future research into TE-mediated genome evolution and could inform novel strategies for crop improvement by leveraging the genetic variation generated by these dynamic elements.
Read the full paper: Huang, Y., Sahu, S.K. and Liu, X. (2024) Deciphering recent transposition patterns in plants through comparison of 811 genome assemblies. Plant Biotechnol. J., https://doi.org/10.1111/pbi.14570