Decoding the Genomic and Cis-Regulatory Basis of Plastic C3-C4 Photosynthesis
Our recent study reveals how Eleocharis baldwinii switches between C3 and C4 photosynthesis through environmentally-triggered, cell-specialized cis-regulatory elements.
We are pleased to announce the publication of “Genomic and Cis-Regulatory Basis of a Plastic $C_3$-$C_4$ Photosynthesis in Eleocharis baldwinii” in Advanced Science. This study, a collaboration between XinLab and Huazhong Agricultural University, utilizes single-cell multiomics to decipher the photosynthetic plasticity of this unique sedge.
The Mystery of Photosynthetic Plasticity
$C_4$ photosynthesis is a major evolutionary trait for efficient carbon fixation. Eleocharis baldwinii exhibits remarkable plasticity, displaying $C_3$-like traits underwater and $C_4$-like traits on land. This environmental adaptability makes it an ideal model to study the origin and regulation of the $C_4$ pathway.
Key Breakthroughs
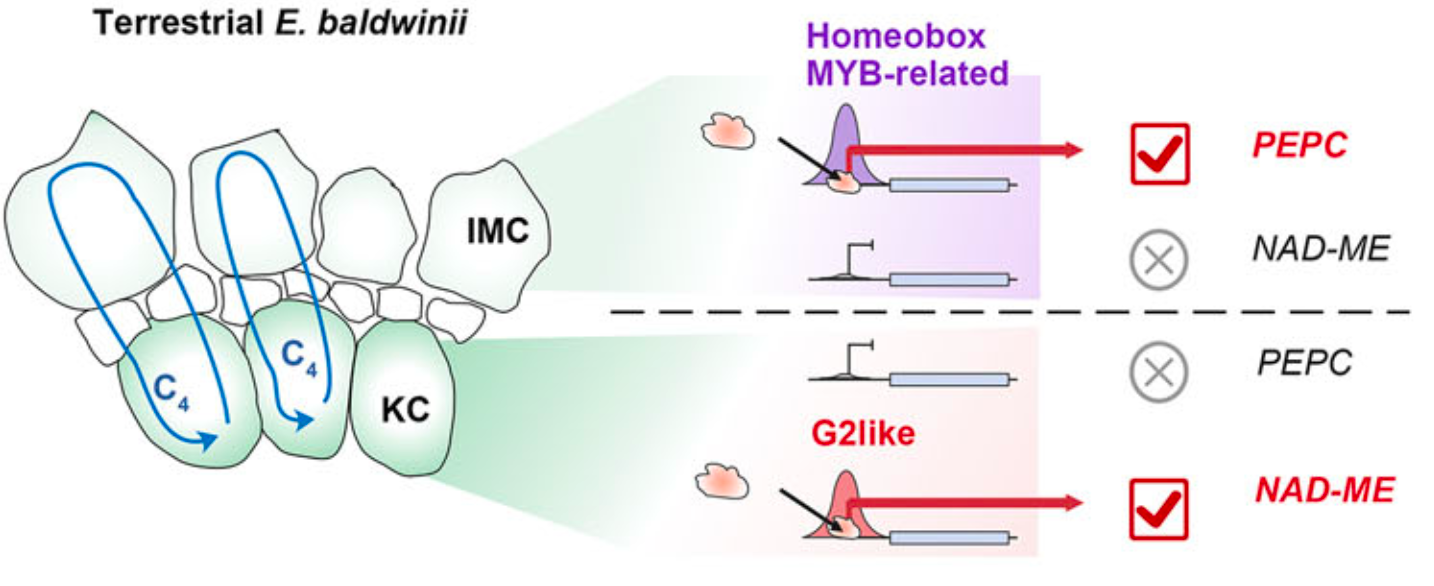
- Gap-free Allopolyploid Genome: We assembled a high-quality, gap-free genome (708.9 Mb) for E. baldwinii, detecting significant subgenome dominance influencing the expression of $C_4$ enzyme genes across different cell types.
- Single-Cell Multiomic Atlas: By applying snRNA-seq and snATAC-seq, we reconstructed a cellular expression and chromatin accessibility atlas. We found that the transition to $C_4$ involves highly specialized gene expression in bundle sheath (BS) and mesophyll (M) cells.
-
Environmentally-Triggered Switches: The formation of the $C_4$ pathway in E. baldwinii is driven by a few specific cis-regulatory elements that are triggered by land-based environmental signals.
- Comparative Evolutionary Insights: Comparisons with maize suggest that while regulatory families for Kranz anatomy are conserved, the specific cis-regulators for $C_4$ metabolism vary, indicating diverse evolutionary paths toward $C_4$ efficiency.
Significance
This research provides a foundational genomic resource for studying photosynthetic evolution. It highlights the potential for enhancing crop yields by manipulating a small number of environmentally-responsive regulatory elements to introduce $C_4$ traits into $C_3$ crops.
Collaborative Impact
This project reflects a deep synergy between plant physiology and advanced genomics. The XinLab team (including Xin Liu and Chuan Chen) led the comprehensive multiomic data integration and regulatory analysis, helping to unravel the complex molecular landscape of photosynthetic adaptation.
Read the full paper here: https://doi.org/10.1002/advs.202405626