Soybean Transcriptomic Atlas
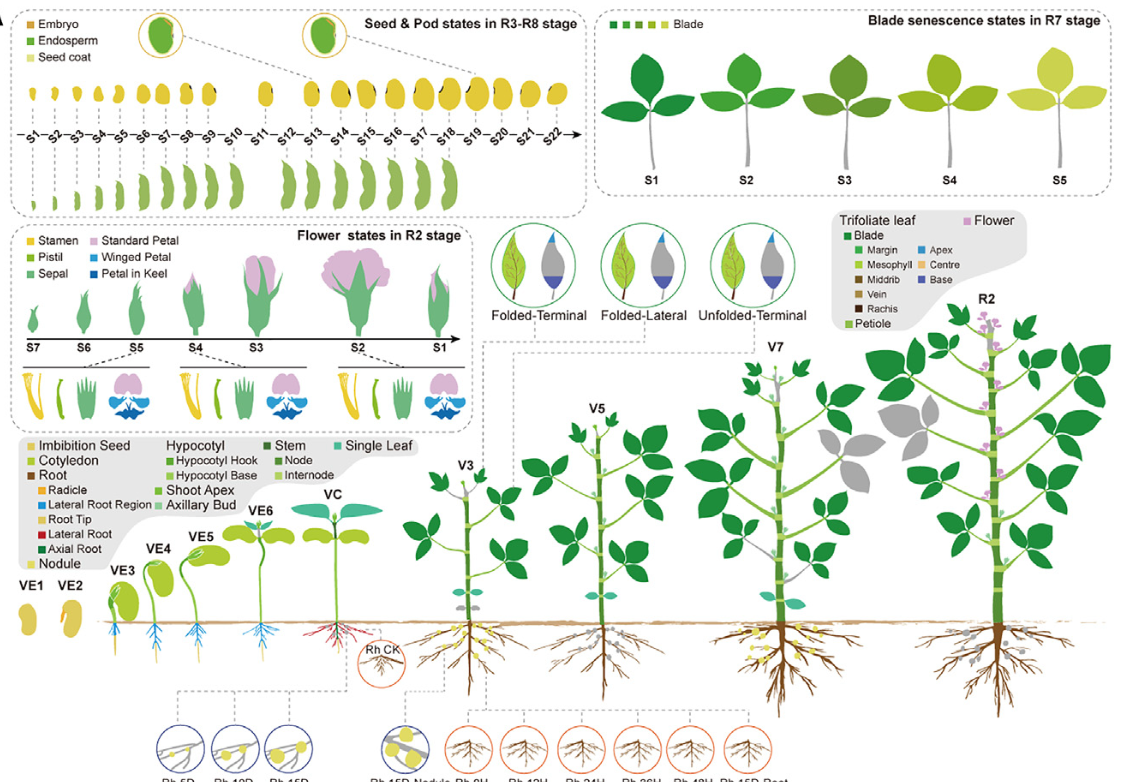
We generated Bulk, Single-Cell, and Spatial Transcriptome Data for Soybean to Unveil Mechanisms Underlying its Organ Development
Release Date: April 7, 2025
Researchers, including teams from XinLab (BGI-Shenzhen/Beijing), in collaboration with the Institute of Genetics and Developmental Biology (Chinese Academy of Sciences) and other institutions, have published a groundbreaking study in the internationally renowned journal Molecular Plant. They have successfully constructed a comprehensive, large-scale integrated transcriptomic atlas of soybean organ development, providing unprecedented insights into the growth regulation of this crucial crop, marking a milestone for future soybean genetic improvement.
💡 Why is this Research Crucial?
Soybeans are a major global source of protein and oil. To meet growing demand and enhance resilience to environmental changes, a deep understanding of the complex genetic networks governing the development of its various organs—such as roots, leaves, flowers, seeds, and symbiotic nitrogen-fixing nodules—is urgently needed. This knowledge is key to breeding higher-yielding, more adaptable varieties.
🔬 An Advanced Multi-Omics Integration Approach
To create this detailed developmental blueprint, the research team utilized the soybean cultivar ‘Zhonghuang 13’ (ZH13) and ingeniously integrated several cutting-edge omics technologies:
- 📊 Bulk RNA Sequencing: Analyzed 314 samples covering the entire soybean life cycle, encompassing 14 organs at different developmental stages, providing a global overview of gene expression.
- 🧬 Single-Nucleus RNA Sequencing (snRNA-seq): Delved into the single-nucleus level to finely resolve cell-type diversity and gene expression profiles within five key organs: root, nodule, shoot apex, leaf, and stem.
- 🗺️ Spatial Transcriptomics (Stereo-seq): Combined high-resolution imaging and sequencing technology to map gene expression in situ within the tissue context of the same five organs, revealing the spatial organization of gene activity and cellular microenvironments.
This multi-dimensional data integration allows researchers to track dynamic gene expression changes across time, space, organs, and cell types with unprecedented resolution.
✨ Key Scientific Discovery Highlights
This powerful integrated atlas enabled several significant scientific discoveries:
- Nodule Symbiotic Nitrogen Fixation Mechanisms:
- Identified novel sugar transporter gene family (
GmPMT) crucial for energy supply during nodule formation. - Discovered key transcription factors (
GmHB1a/b,GmHB21of the HD-ZIP I family) with precise spatiotemporal regulatory roles in nodule vascular bundle development.
- Identified novel sugar transporter gene family (
- Leaf Development & Morphogenesis:
- Revealed the central role of the transcription factor
MYB94and its regulatory network in cuticular wax biosynthesis in young, unfolded leaves.
- Revealed the central role of the transcription factor
- Cell Fate Atlas & Developmental Trajectories:
- Generated detailed cell atlases for key organs, for instance, locating
GmPLTgenes potentially involved in maintaining stem cell characteristics in the root tip. - Distinguished specific cell populations and their functional transitions between early and mature nodule development stages by comparing nodules at different time points, involving processes like hypoxia response, nodulin synthesis, and cell wall remodeling.
- Generated detailed cell atlases for key organs, for instance, locating
- Post-Transcriptional Regulation:
- Identified organ-specific alternative splicing (AS) events, suggesting their potential regulatory roles in organ functional differentiation.
🌐 An Open Resource Treasure Trove for the Community
A core value of this work lies in its openness. Beyond publishing the findings, the team created a publicly accessible, user-friendly online database. All data have been integrated into the SoyOmics platform and a dedicated Soybean Organ Transcriptomic Atlas website. Researchers worldwide can freely access, query, download, and visualize this valuable data resource (including 3D reconstruction models for root and nodule), which will greatly accelerate functional genomics research and related breeding applications in soybean.
🚀 Significance & Future Outlook
This integrated transcriptomic atlas study, co-led by XinLab (Dr. Xin Liu’s team) and the Institute of Genetics and Developmental Biology, CAS (Dr. Zhixi Tian’s team), provides an invaluable resource for the global soybean research community. It not only deepens our understanding of the complexity of plant organogenesis but also paves the way for precisely identifying key genes determining important agronomic traits, thereby accelerating the breeding of high-yield, high-quality, and resilient soybean varieties.
💡 Some of my personal comments
Fifteen or sixteen years ago, I began my journey into the field of genomics, with my first project focusing on soybean resequencing. At that time, we conducted whole-genome resequencing on 17 wild soybeans and 14 cultivated soybeans, marking the first study on soybean population diversity at the whole-genome level. After the publication of this project, Professor Tian Zhixi, who was then working in Dr. Ma Jianxin’s lab in the United States, reached out to us for the data. He conducted research on the identification and analysis of repetitive sequences in the soybean genome, which was later published as well.
Professor Tian subsequently returned to China, joining the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, where he continued soybean-related research and published a series of high-quality papers, including studies on the soybean pan-genome. Around 2021, following the establishment of our joint research center at the Genetics Institute, I had the opportunity to meet Professor Tian in person for the first time (our previous interactions had been limited to emails, despite co-authoring research articles). We quickly decided to leverage the platform we had built in Beijing to collaborate on soybean spatiotemporal genomics research.
Over the past few years, we divided tasks and worked collaboratively, efficiently completing the study described in this paper. Throughout the process, I gained a deep appreciation for Professor Tian and his team’s outstanding scientific approach, which has greatly benefited both myself and Chen Chuan.
📄 Publication & Data Access Details
- Journal: Molecular Plant
- Title: A large-scale integrated transcriptomic atlas for soybean organ development
- Authors: Fan J., Shen Y., Chen C., Chen X., et al.
- Volume/Pages: 18, 669-689 (2025)
- DOI: 🔗 https://doi.org/10.1016/j.molp.2025.02.003
- Database Links:
- 🌐 SoyOmics Integrated Database: https://ngdc.cncb.ac.cn/soyomics/transcriptome
- 🗺️ Soybean Organ Transcriptomic Atlas (incl. Spatial Data & 3D Models): https://db.cngb.org/stomics/soybean
- Funding: This research was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Taishan Scholars Program, and the Xplorer Prize, among others.
- (Optional) Contact: [Insert your lab’s contact email, website, or other information here]