Single‐Cell Multiome Reveals Root Hair‐Specific Responses to Salt Stress in Non‐Heading Chinese Cabbage
Our recent paper introduced findings on mechanisms underlying salt-tolerance in non-heading Chinese cabbage using single-cell and spatial transcriptome sequencing.
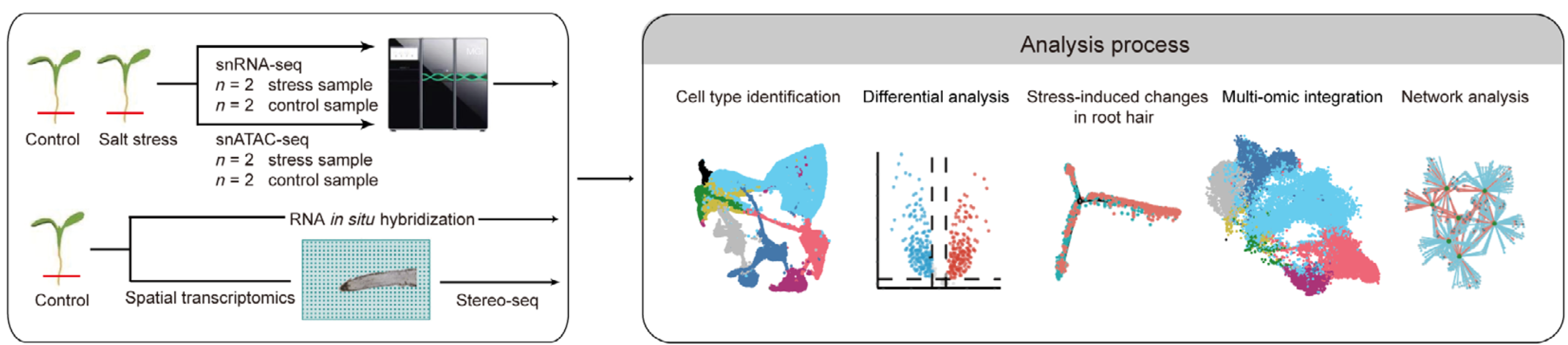
We are excited to announce that our latest research article, “Single‐cell multiome reveals root hair‐specific responses to salt stress,” has been published in New Phytologist (DOI: 10.1111/nph.70160). This work marks a significant advance in our understanding of how non‐heading Chinese cabbage (NHCC) roots adapt at the cellular and molecular levels to salt stress—a growing challenge for sustainable agriculture.
Background
Soil salinization is a critical issue driven by environmental degradation and unsustainable cultivation practices. Salt stress affects plants by:
- Inhibiting seed germination and root development
- Reducing crop yield through detrimental physiological impacts
For NHCC—a vital leafy vegetable crop—the root system is central to detecting and responding to salinity. However, the cell type–specific regulatory mechanisms underlying this response have largely remained unexplored. To address this gap, our study leverages cutting-edge single-cell multiomic technologies to create the first comprehensive single-cell atlas of NHCC root tips under both normal and salt-stress conditions.
Methodology
Our approach combined several advanced techniques to dissect the complexity of root responses:
- Single-Nucleus RNA Sequencing (snRNA-seq) and ATAC Sequencing (snATAC-seq):
- Nuclei were isolated from 5-mm root tips of NHCC plants grown under control conditions and after a 12-hour treatment with 150 mM NaCl.
- This dual approach allowed us to simultaneously profile gene expression and chromatin accessibility, revealing distinct cell populations and regulatory elements.
- Spatial Transcriptomics (Stereo-seq):
- Root tip sections were processed with Stereo-seq, capturing spatial expression data at “cell-sized” resolution.
- This enabled us to map the anatomical distribution of cell type–specific markers and correlate transcriptomic data with tissue structure.
- RNA In Situ Hybridization and Histochemical Staining:
- These validation experiments confirmed the subcellular localization of important genes.
- Histochemical stains (e.g., Perls and DAB) were used to assess iron distribution in root tissues.
- Virus-Induced Gene Silencing (VIGS):
- We silenced the iron transporter gene BcIRT2 to decipher its role under salt stress.
- Plants with reduced BcIRT2 expression exhibited lower iron accumulation and stress symptoms (e.g., leaf chlorosis), underscoring its pivotal role.
Key Findings
- Disrupted Root Hair Differentiation
- Normal vs. Salt Stress Conditions: Under salt stress, the developmental trajectory of root hair cells was markedly altered. Many cells were arrested in an undifferentiated state, preventing the proper transcriptional program required for full maturation
- Suppression of Stress-Response Genes: The focus on single-cell transcriptomics revealed that key stress-responsive genes were not activated in the root hair clusters, directly impairing nutrient uptake and environmental sensing abilities.
- Impaired Iron Uptake Linked to BcIRT2
- Downregulation of BcIRT2: We discovered that BcIRT2, the NHCC ortholog of an established Arabidopsis iron transporter, was significantly downregulated under salt stress.
- Physiological Impact: Reduced BcIRT2 levels correlated with lower iron content in roots. VIGS experiments confirmed that silencing BcIRT2 produced chlorotic leaves and increased salt sensitivity, highlighting the gene’s importance in sustaining iron homeostasis.
- Cell Type–Specific Regulatory Networks
- Integrated Multiomic Analysis: By combining snRNA-seq, snATAC-seq, and spatial transcriptomics, we constructed a high-resolution atlas that delineates the distinct regulatory circuits of individual cell types in the NHCC root tip.
- Regulatory Insights for Crop Improvement: Our study identified key transcription factors and signaling pathways that govern root hair differentiation and stress response. These findings provide molecular targets for future breeding programs aimed at enhancing salt tolerance.
Discussion & Impact
Our research provides a comprehensive view of the cellular reprogramming that occurs in NHCC roots under salt stress. The arrest in root hair differentiation and the consequent impairment in iron uptake are critical factors that could explain stress-induced yield losses. Importantly, this work:
- Deepens our fundamental understanding of plant abiotic stress responses at the single-cell level.
- Offers valuable molecular insights that can drive the development of salt-tolerant cultivars.
- Provides a resource-rich multiomic atlas that will support future studies on plant stress physiology and crop improvement.
Acknowledgements
This research was a collaborative effort involving contributions from multiple institutions:
- State Key Laboratory of North China Crop Improvement and Regulation, Hebei Agricultural University
- BGI Research, Beijing (as well as XinLab, www.xgenome.cn)
- College of Life Sciences, University of Chinese Academy of Sciences, Beijing
- Suzhou Academy of Agricultural Sciences, Suzhou
- Glbizzia Biosciences, Beijing
Co-first authors: Qing Liu, Jingmin Kang, Lin Du, and Zhaokun Liu Additional key team members include Hao Liang, Kailai Wang, Haijiao He, Xiaonan Zhang, Qifan Wang, Yiguo Hong, Qi Cheng, Xin Liu, Wei Ma, and Jianjun Zhao. We also gratefully acknowledge the support from:
- National Natural Science Foundation of China (Grant Nos. 32330096; 32202505; U24A20417)
- Hebei Natural Science Foundation (Grant Nos. C2023204308; C2024204246)
- Hebei S&T Bureau (2023HBQZYCSB009)
- Science Foundation of the Ministry of Education of China (MoE, 902/1123401)
Conclusion
The publication of our paper in New Phytologist marks an important milestone in plant genomics research. By unveiling the single-cell landscape of NHCC root responses to salt stress, we have not only expanded our understanding of plant stress physiology but also identified potential targets to enhance crop resilience in saline environments. We are enthusiastic about the future implications of these findings for precision breeding and sustainable agriculture.
Read the full paper here: https://doi.org/10.1111/nph.70160